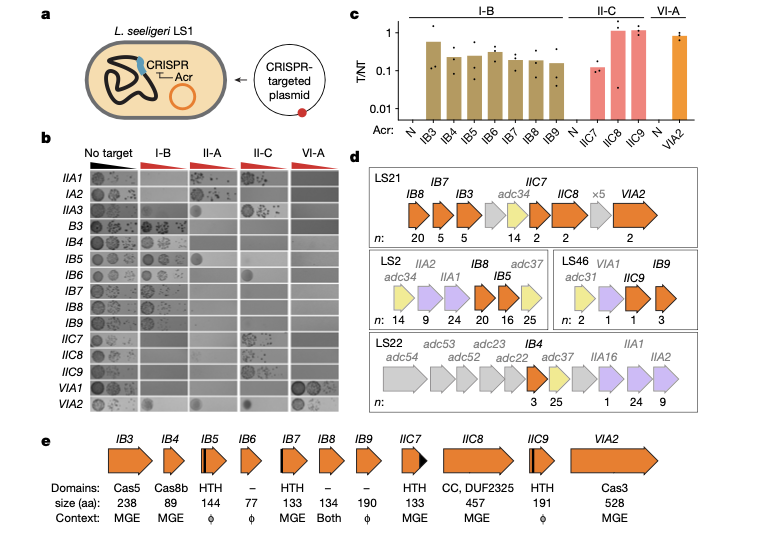
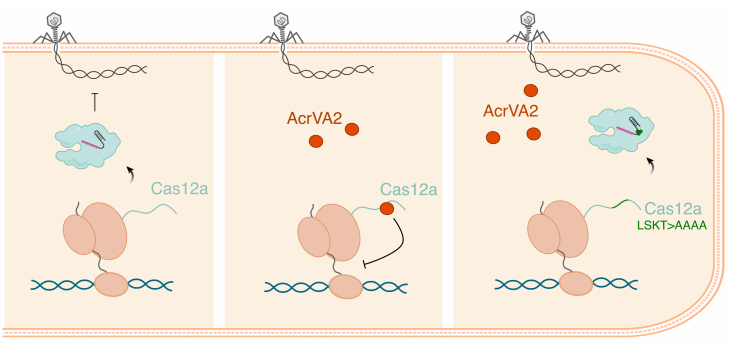
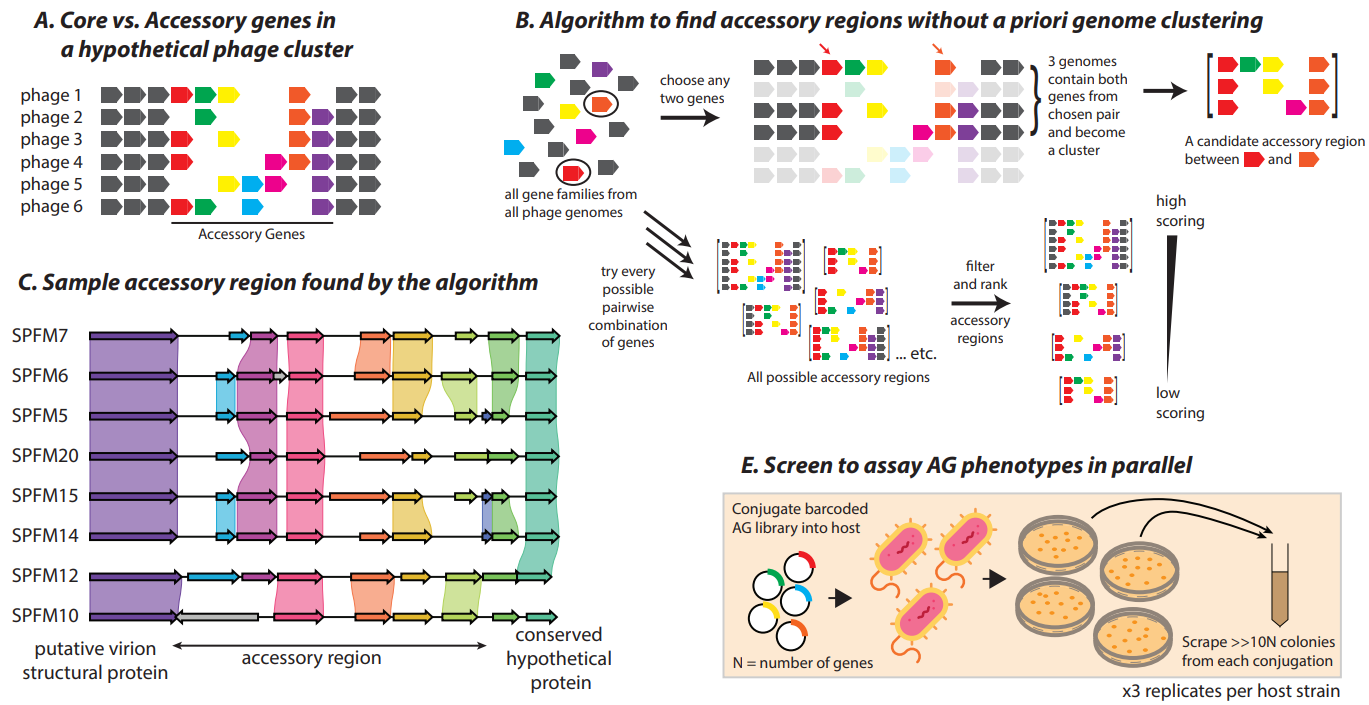
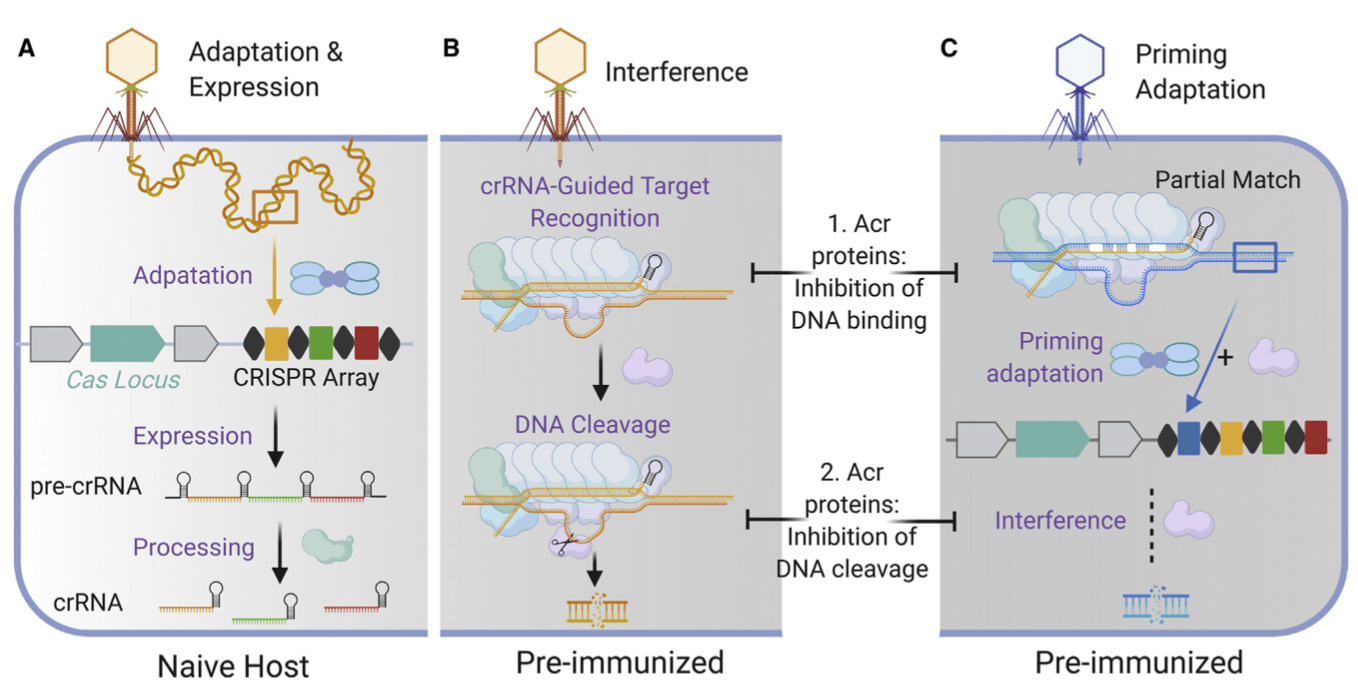
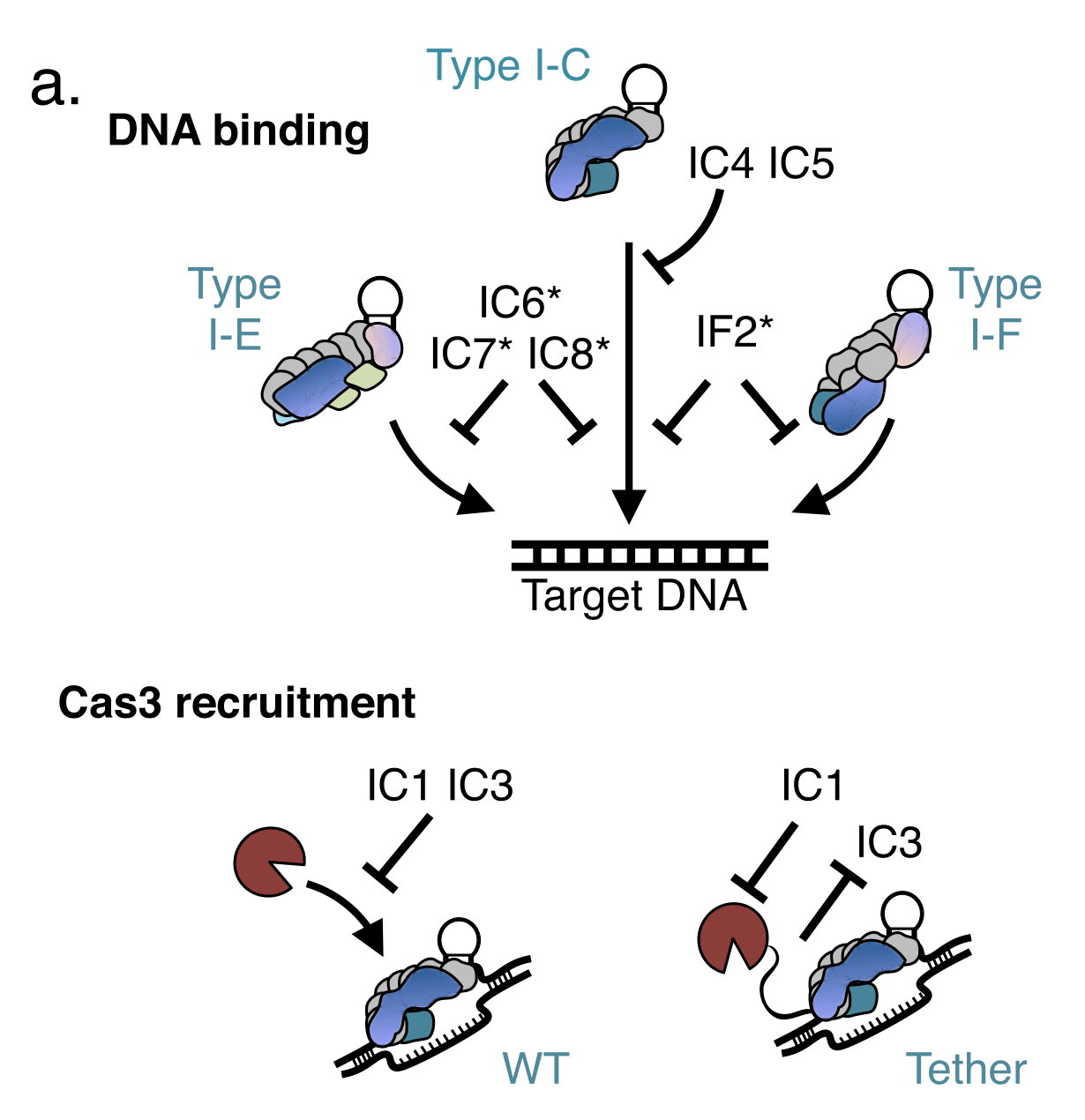
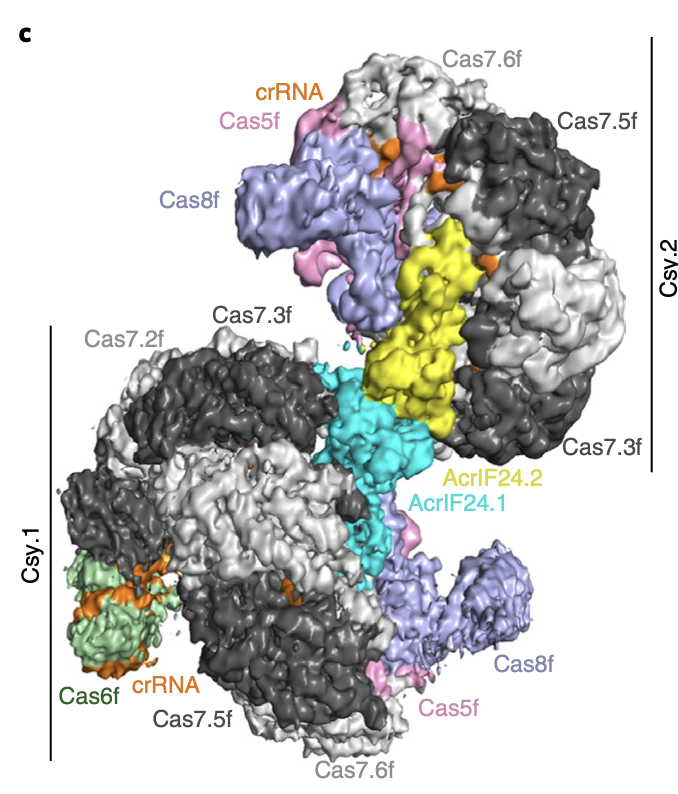
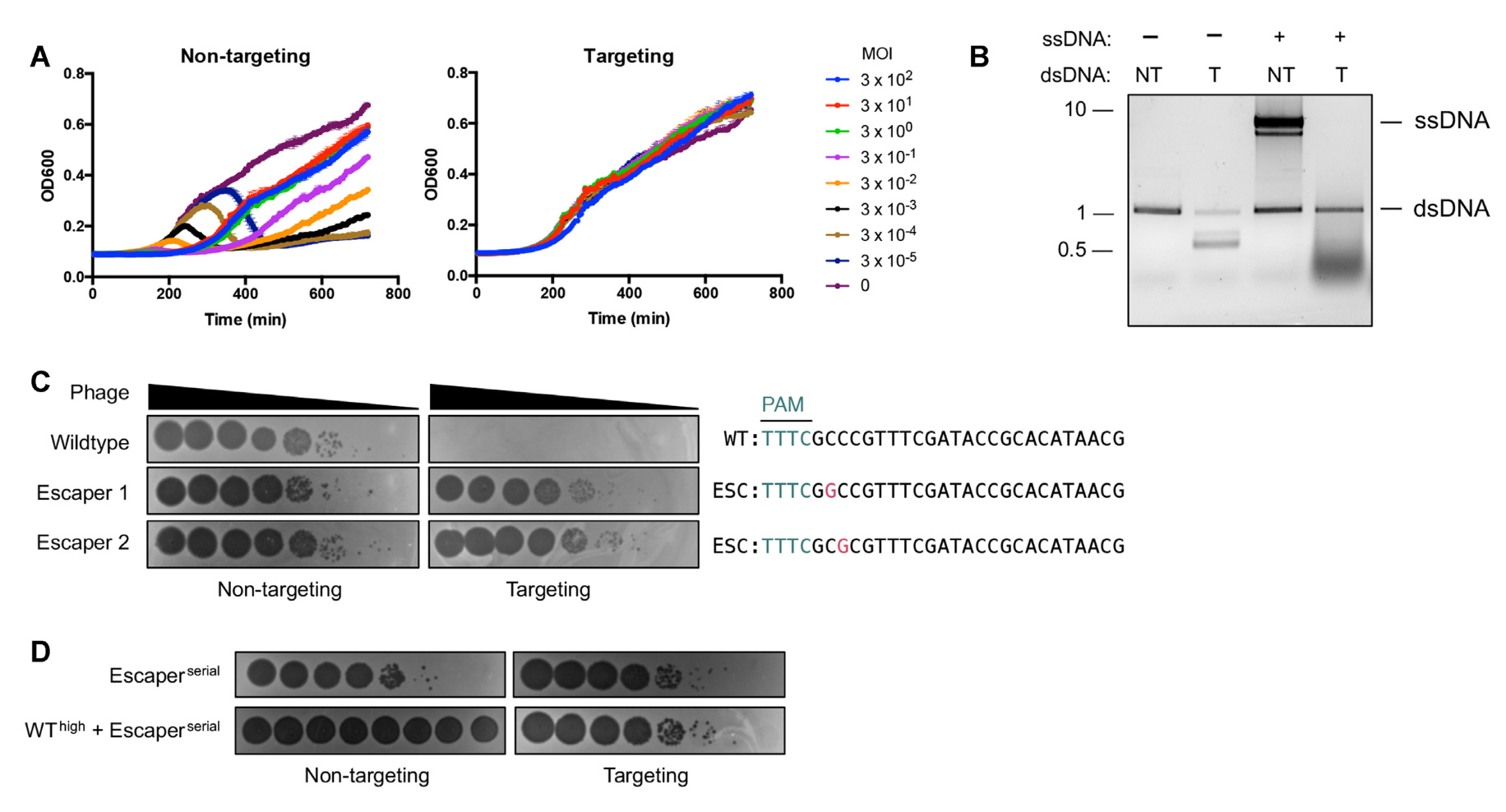
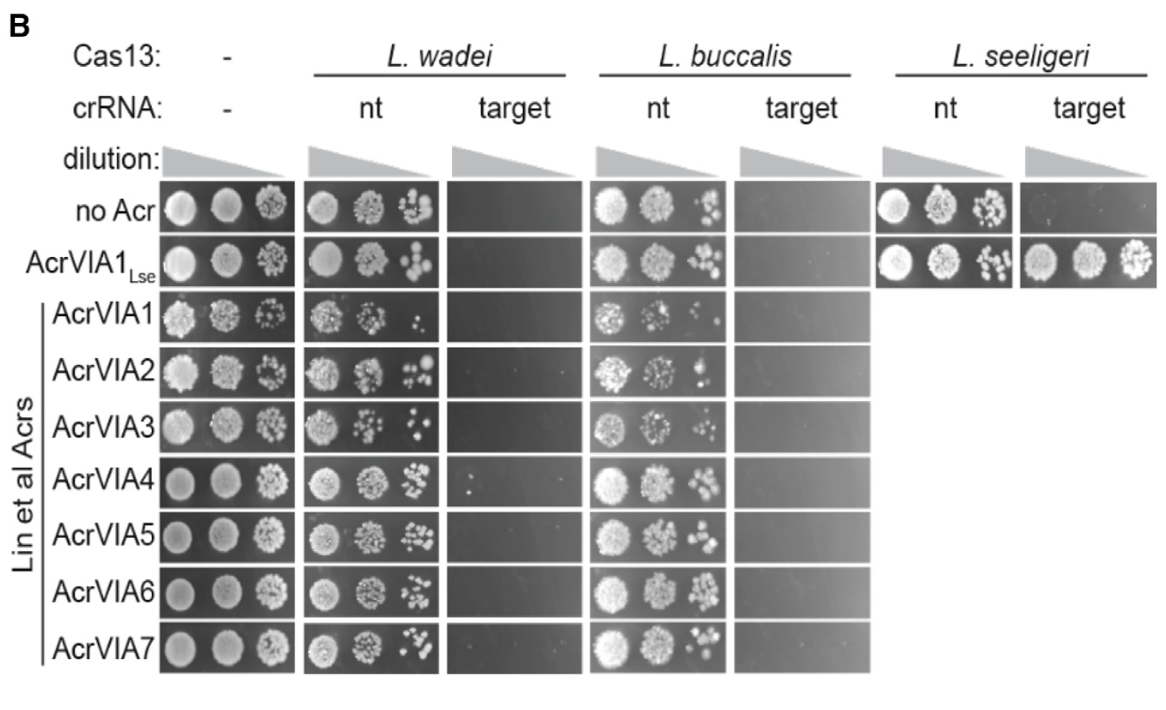
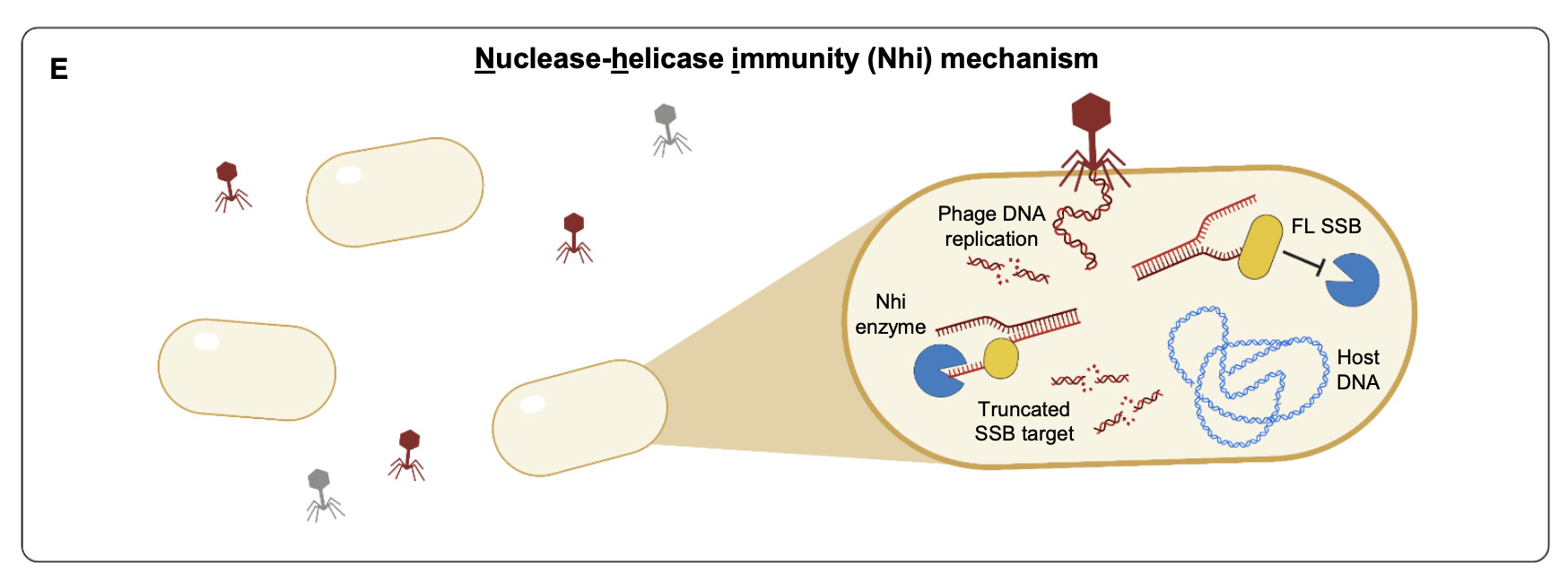
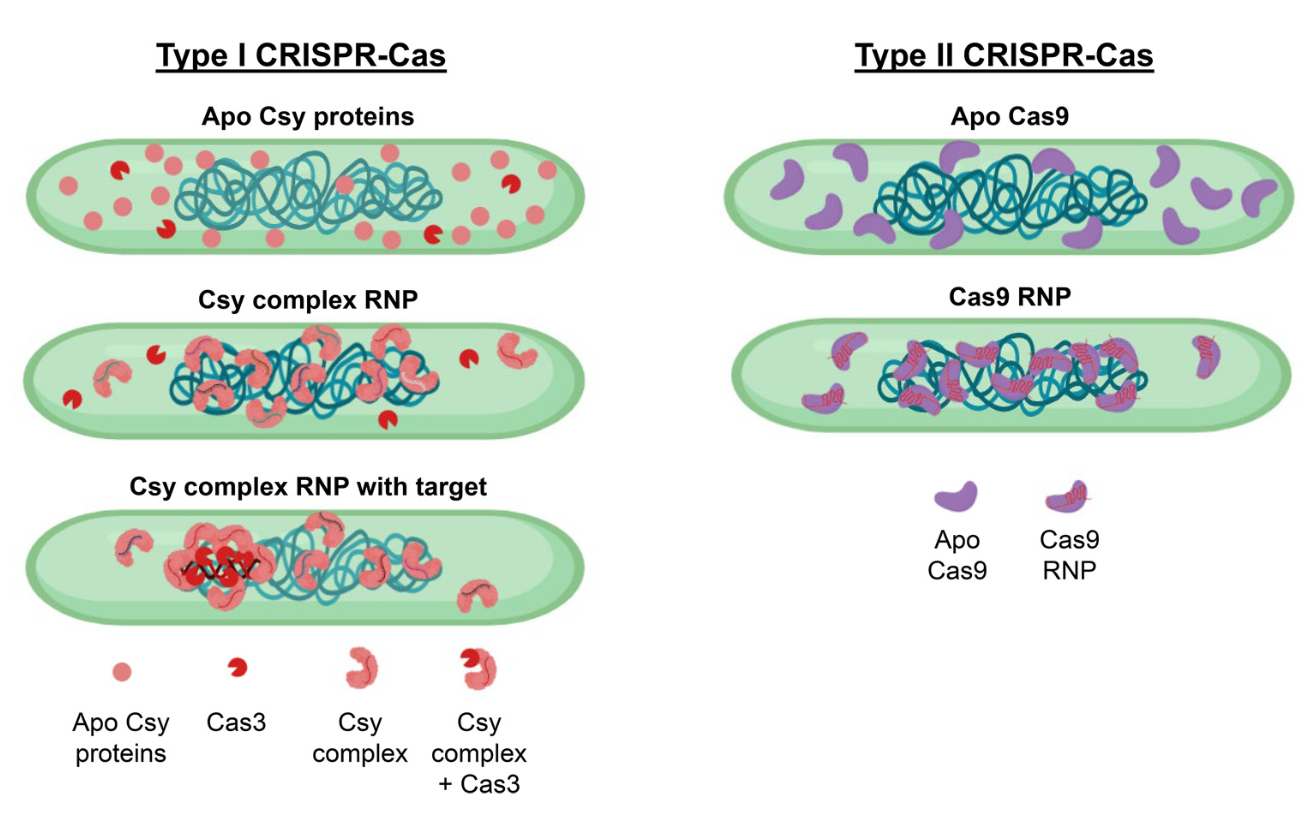
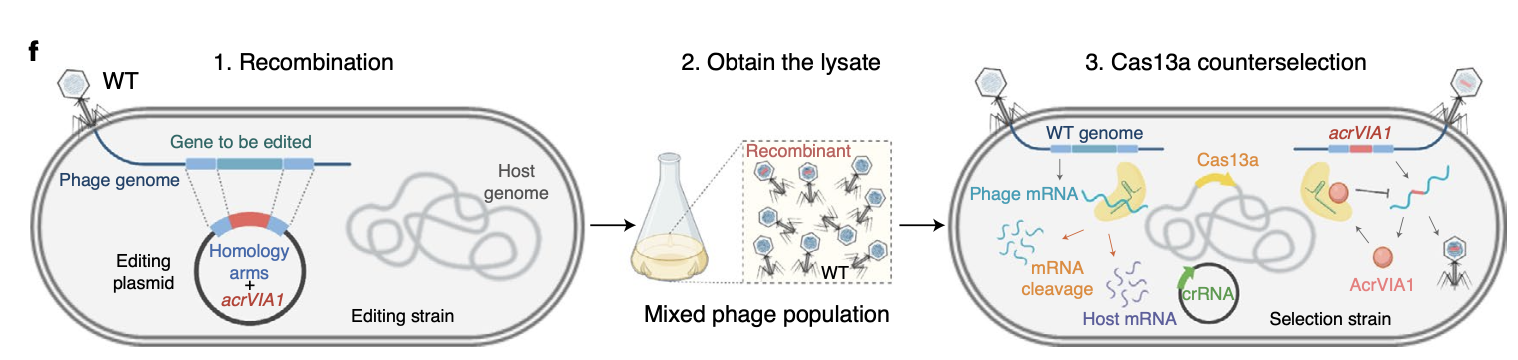
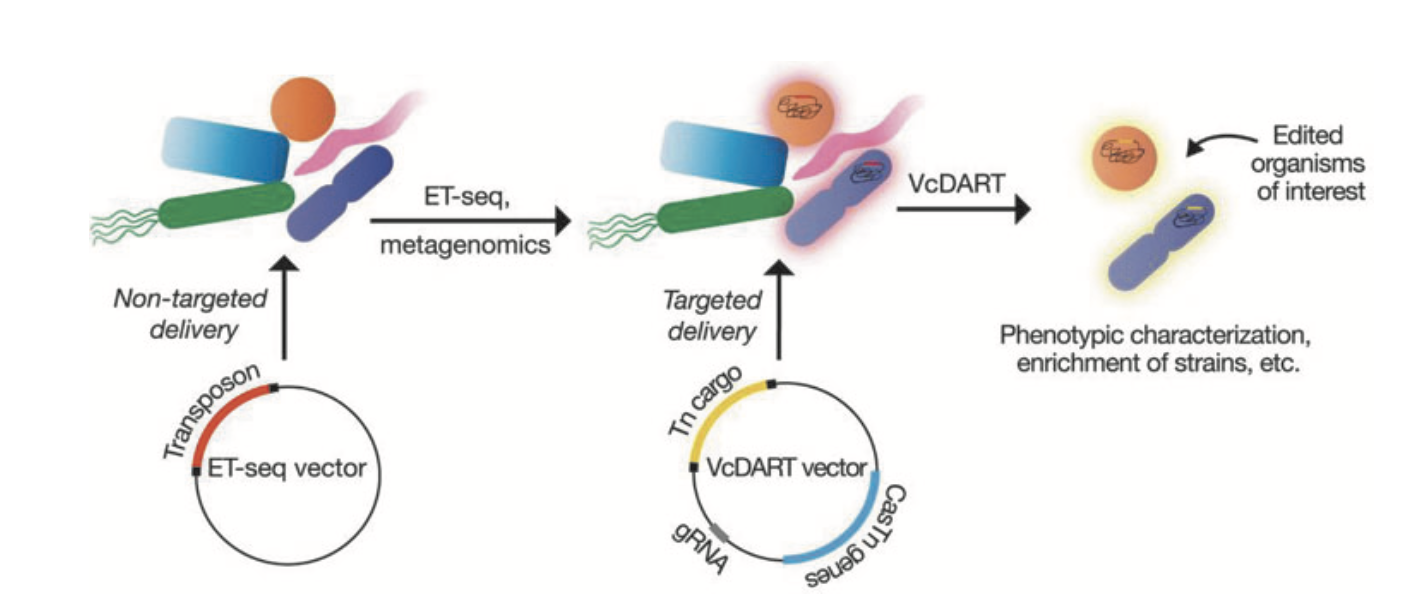
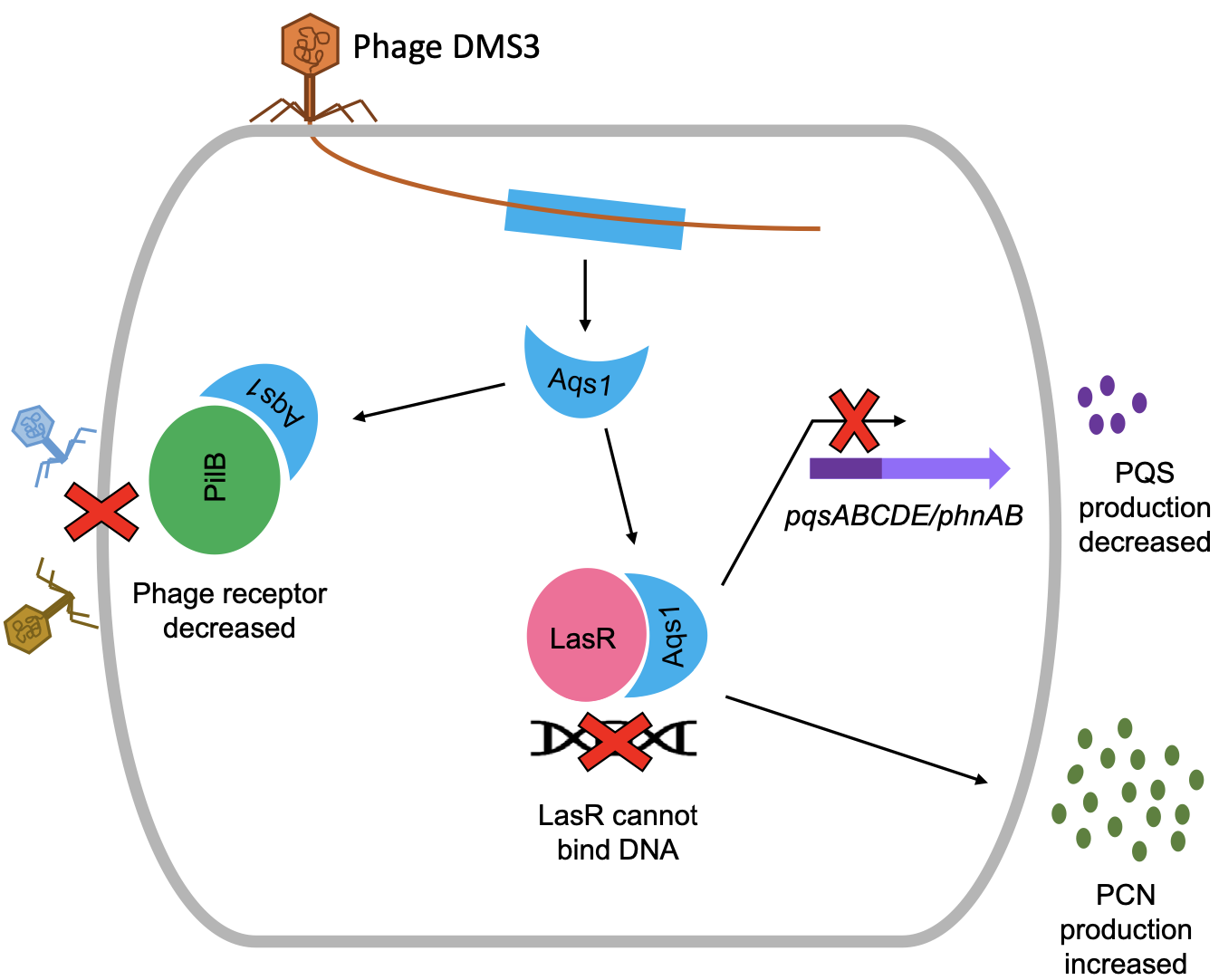
Activation of bacterial programmed cell death by phage inhibitors of host immunity
Silas S, Carion H, Makarova KS, Laderman ES, Todeschini T, Kumar P, Johnson M, Bocek M, Nobrega FL, Koonin EV, Bondy-Denomy J
Molecular Cell,
2025
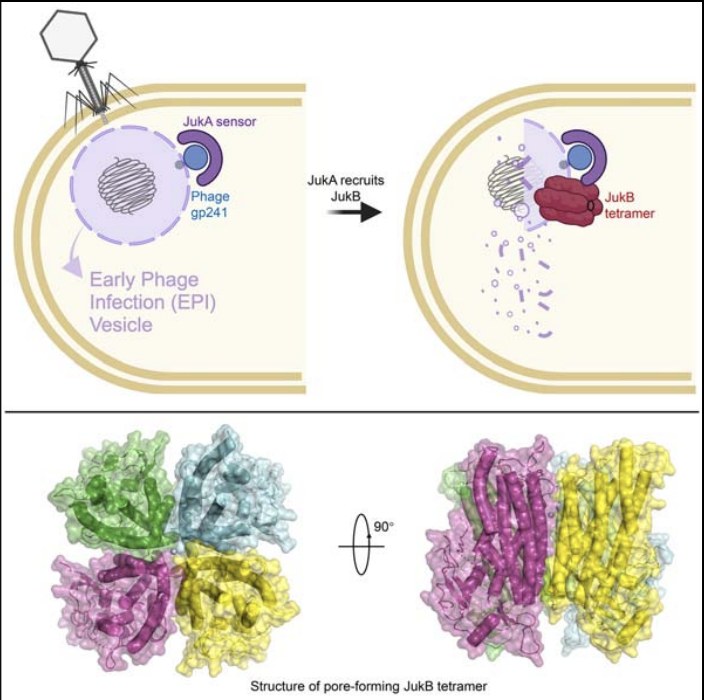
Jumbo phage killer immune system targets early infection of nucleus-forming phages
Li Y, Guan L, Becher I, Makarova KS, Cao X, Hareendranath S, Guan J, Stein F, Yang S, Boergel A, Lapouge K, Remans K, Agard D, Savitski M, Typas A, Koonin EV, Feng Y, Bondy-Denomy J
Cell,
2025
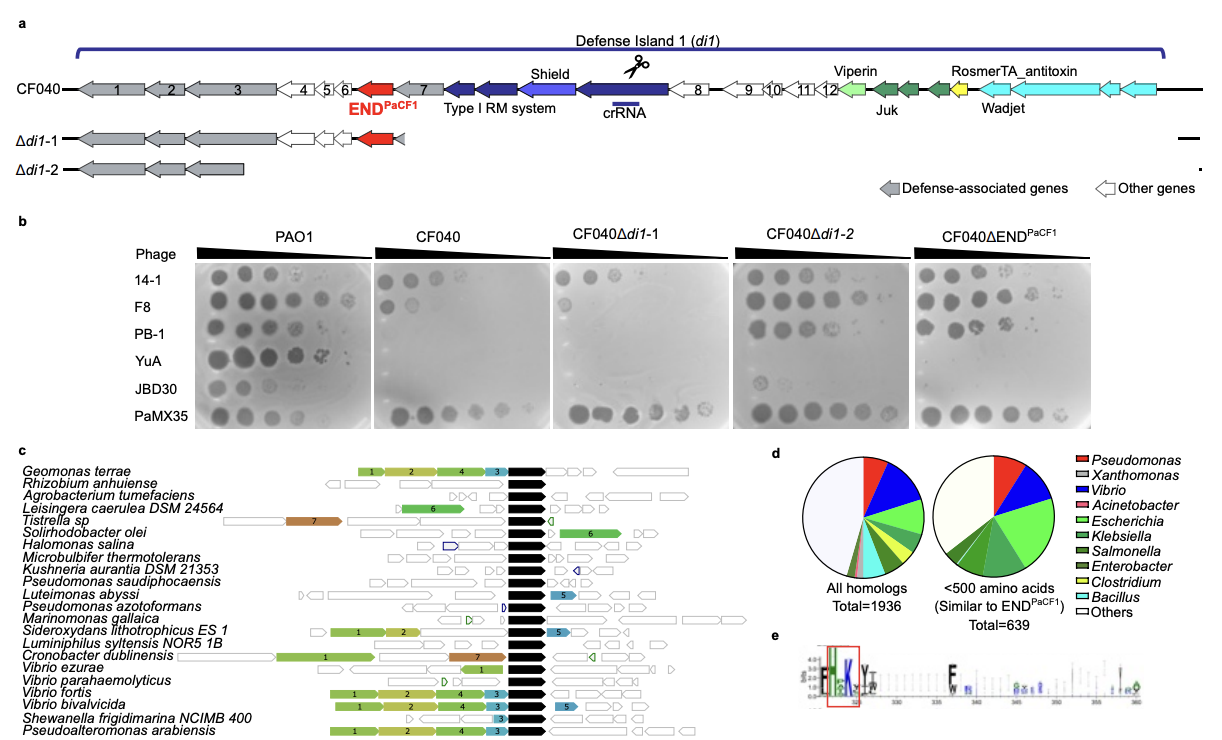
END nucleases: Antiphage defense systems targeting multiple hypermodified phage genomes
Yee W-X, Lee Y-J, Klein TA, Wirganowicz A, Gabagat AE, Csörgő B, Makarova KS, Koonin EV, Weigele PR, Bondy-Denomy J
bioRxiv,
2025
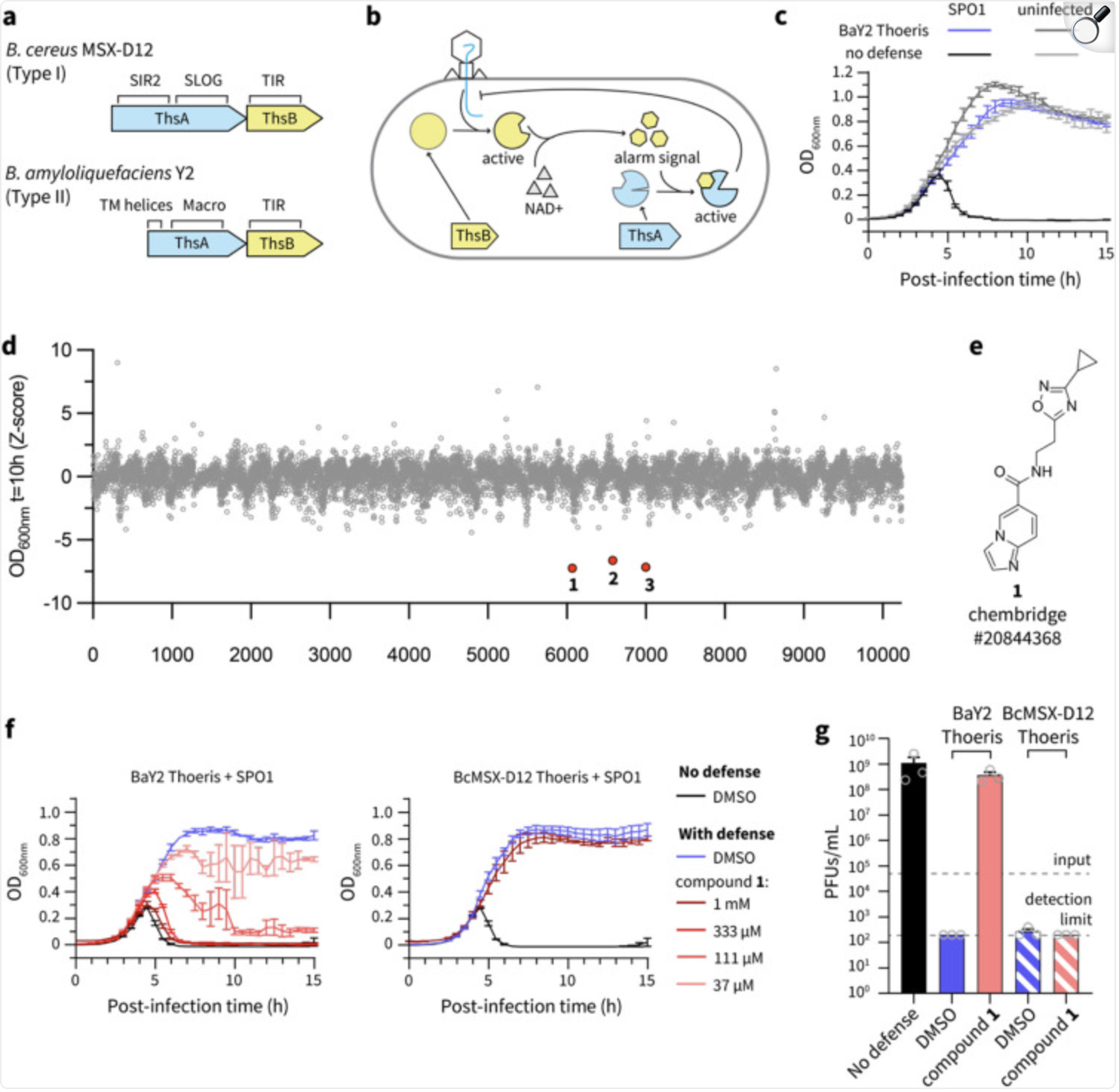
Chemical inhibition of a bacterial immune system
Zang Z, Duncan OK, Sabonis D, Shi Y, Miraj G, Fedorova I, Le S, Deng J, Zhu Y, Cai Y, Zhang C, Arya G, Duerkop BA, Liang H, Bondy-Denomy J, Ve T, Tamulaitiene G, Gerdt JP
bioRxiv,
2025
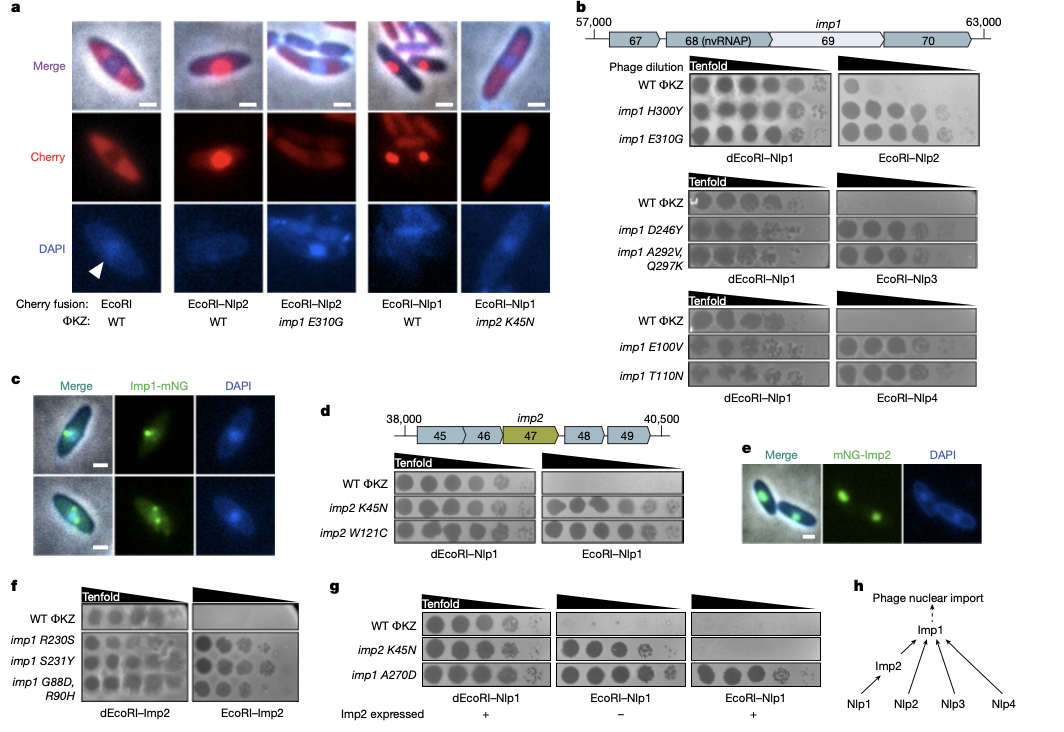
Multi-interface licensing of protein import into a phage nucleus
Kokontis C, Klein TA, Silas S, Bondy-Denomy J
Nature,
2025
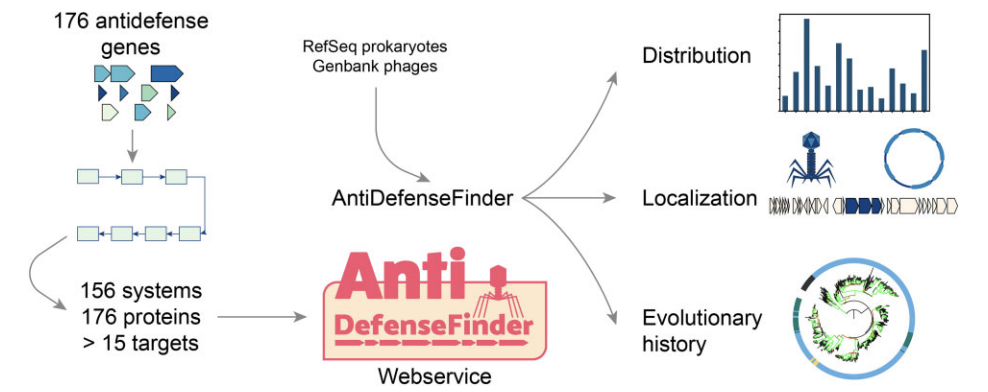
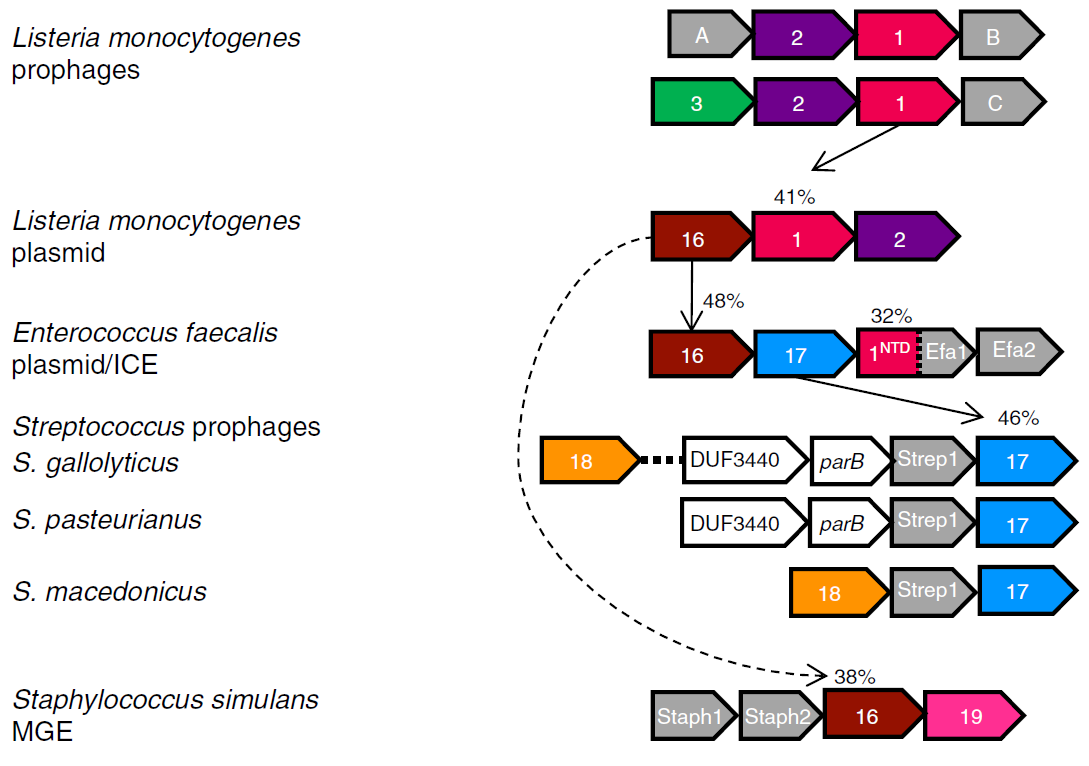
Exploring the diversity of anti-defense systems across prokaryotes, phages and mobile genetic elements
Tesson F, Huiting E, Wei L, Ren J, Johnson M, Planel R, Cury J, Feng Y, Bondy-Denomy J, Bernheim A
Nucleic Acids Research,
2025
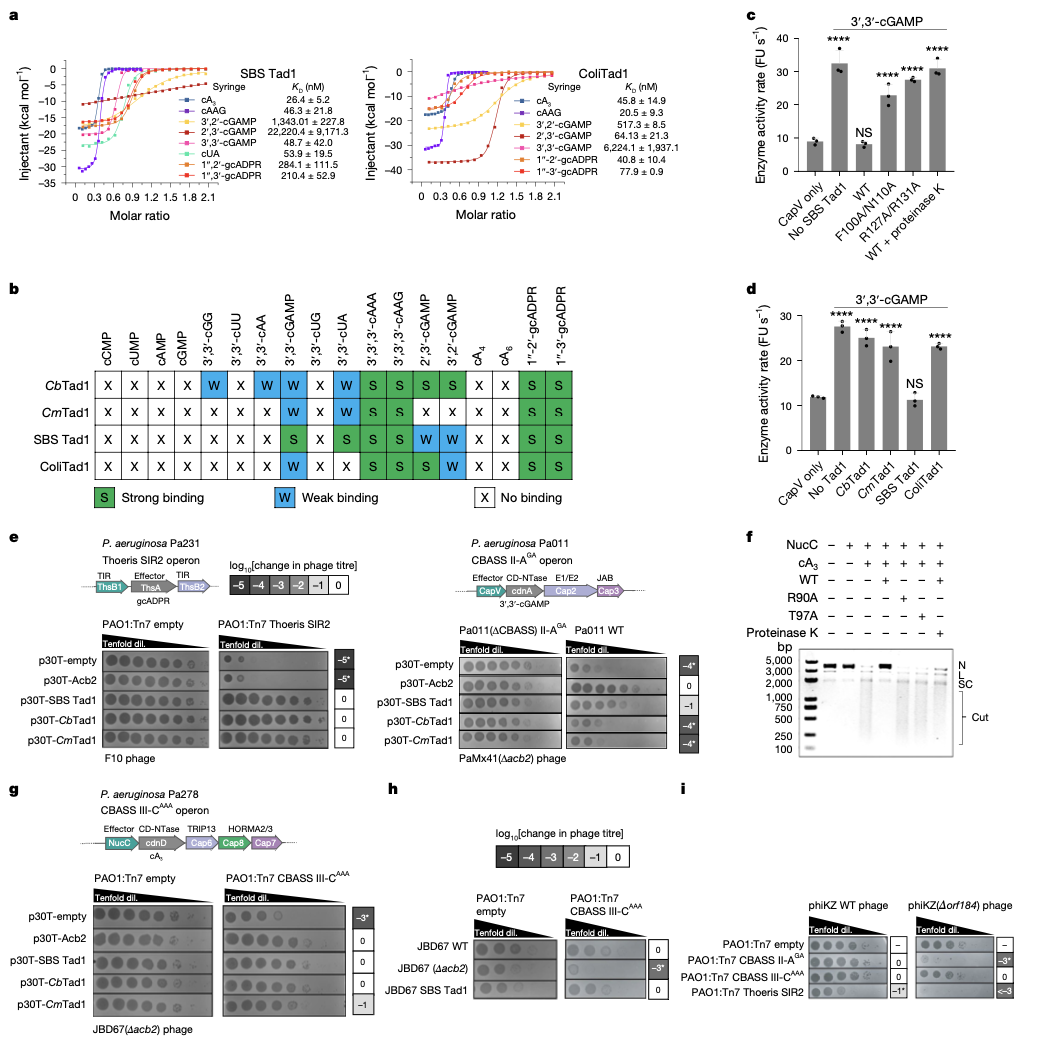
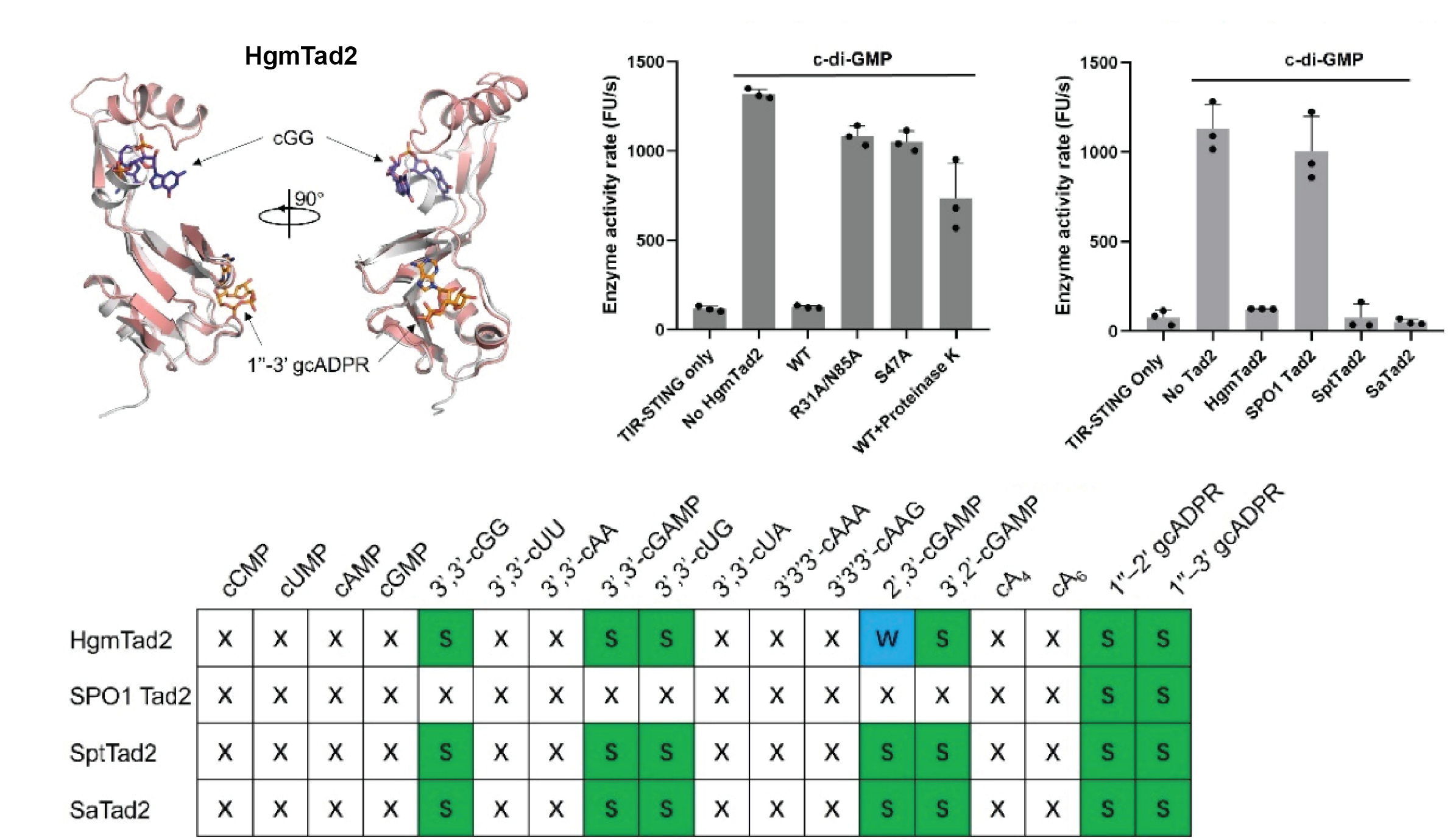
Single phage proteins sequester signals from TIR and cGAS-like enzymes
Li D, Xiao Y, Fedorova I, Xiong W, Wang Y, Liu X, Huiting E, Ren J, Gao Z, Zhao X, Cao X, Zhang Y, Bondy-Denomy J, Feng Y
Nature,
2024
Diverse viral cas genes antagonize CRISPR immunity
Katz MA, Sawyer EM, Oriolt L, Kozlova A, Williams MC, Margolis SR, Johnson M, Bondy-Denomy J, Meeske AJ
Nature,
2024
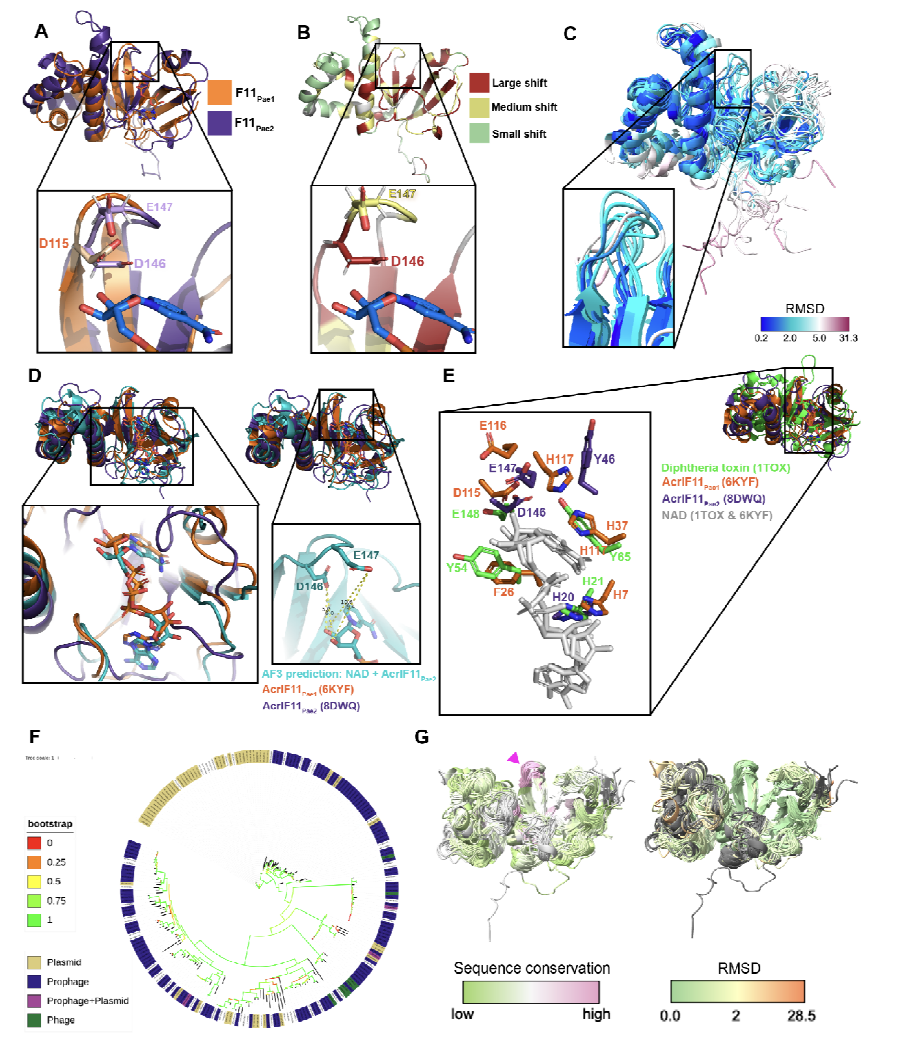
AcrIF11 is a potent CRISPR-specific ADP-ribosyltransferase encoded by phage and plasmid
Chen DF, Roe LT, Li Y, Borges AL, Zhang JY, Babbar P, Maji S, Stevens MGV, Correy GJ, Diolaiti ME, Smith DH, Ashworth A, Stroud RM, Kelly MJS, Bondy-Denomy J, Fraser JS
bioRxiv,
2024
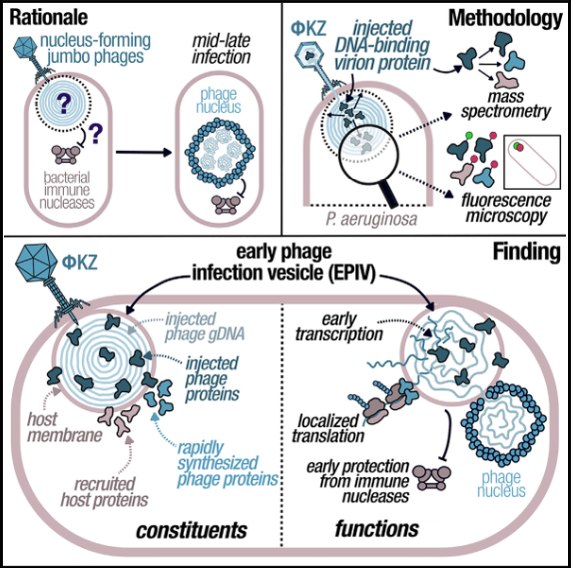
Characterization of a lipid-based jumbo phage compartment as a hub for early phage infection
Mozumdar D, Fossati A, Stevenson E, Guan J, Nieweglowska E, Rao S, Agard D, Swaney DL, Bondy-Denomy J
Cell Host & Microbe,
2024
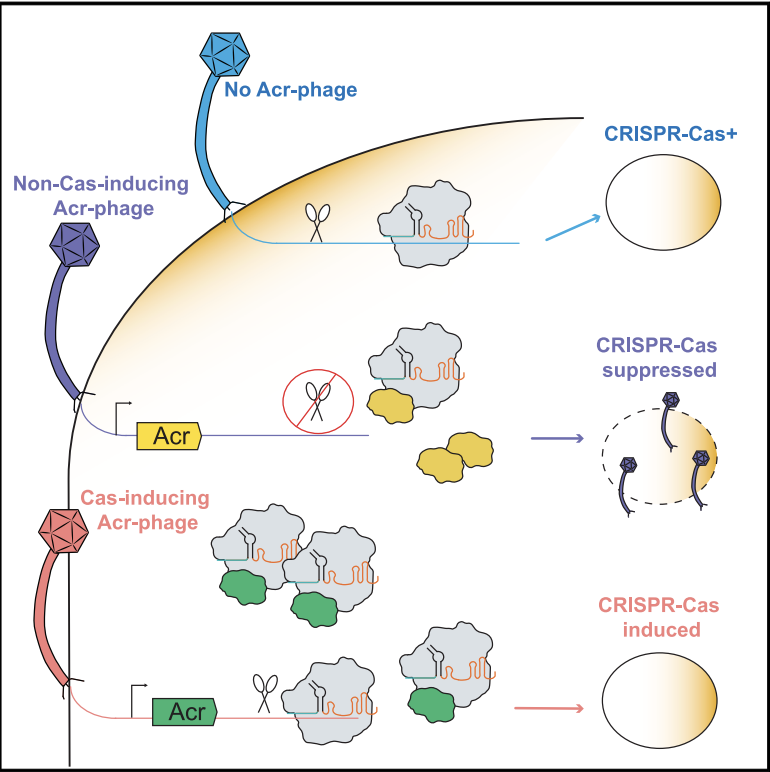
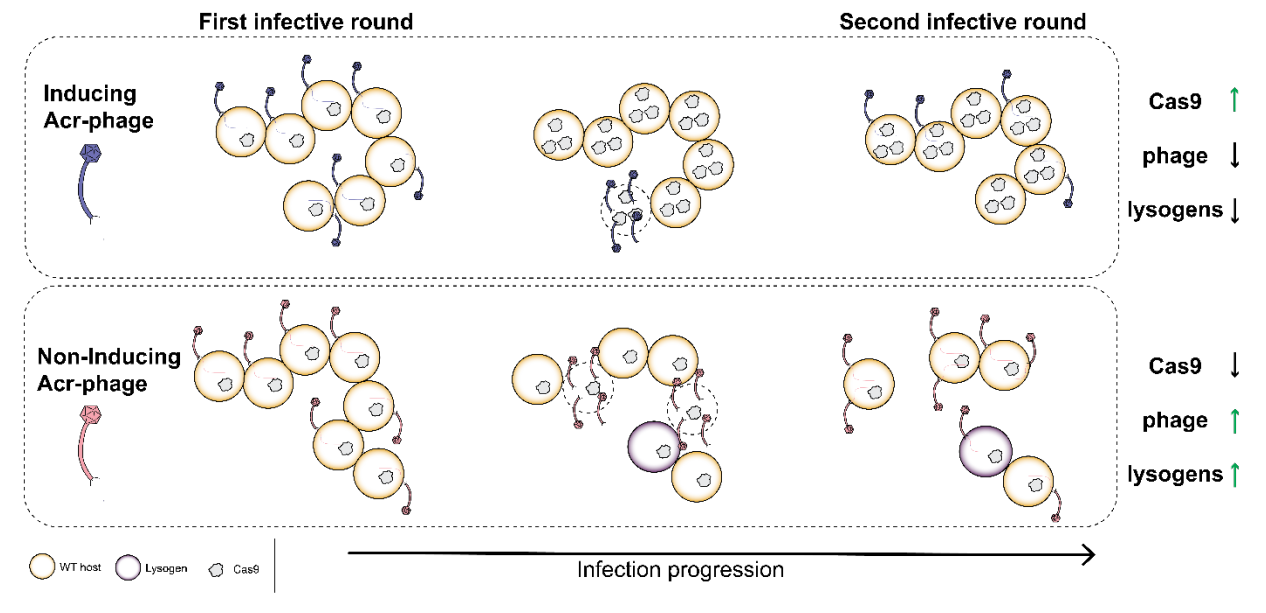
Anti-CRISPR proteins trigger a burst of CRISPR-Cas9 expression that enhances phage defense
Workman RE, Stoltzfus MJ, Keith NC, Euler CW, Bondy-Denomy J, Modell JW.
Cell Reports,
2024
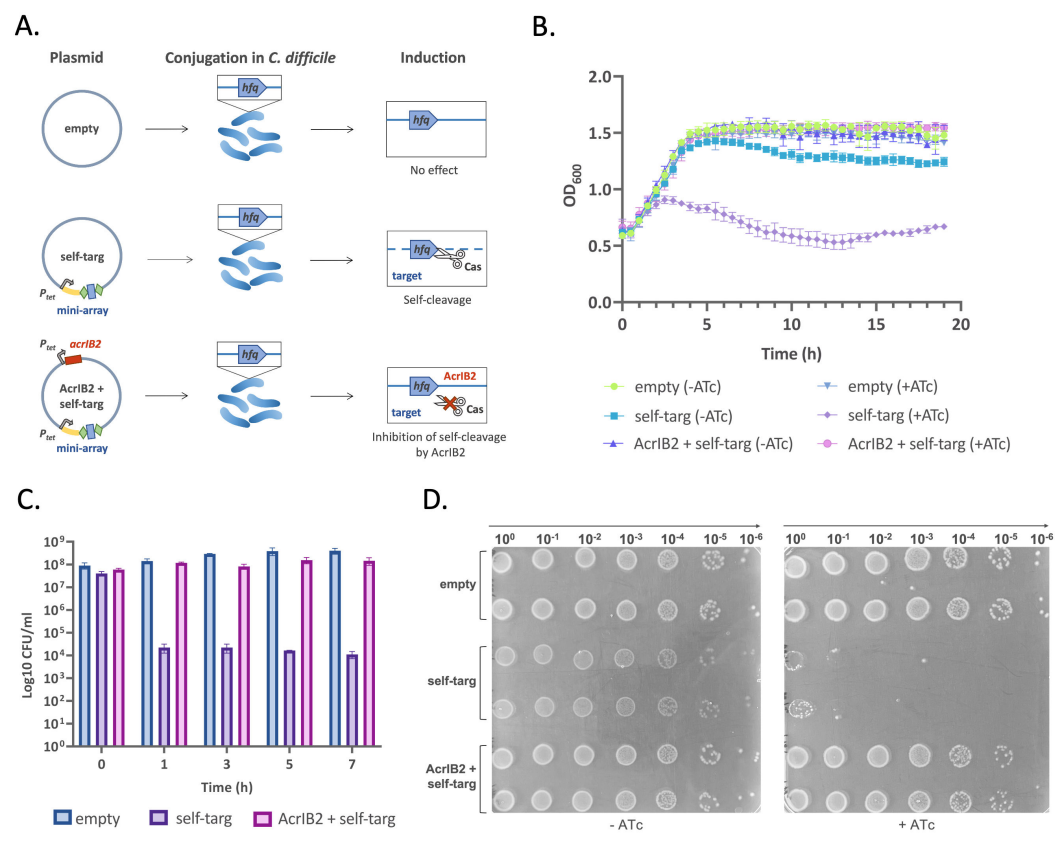
Identification of an anti-CRISPR protein that inhibits the CRISPR-Cas type I-B system in Clostridioides difficile
Polina Muzyukina, Anton Shkaruta, Noemi M. Guzman, Jessica Andreani, Adair L. Borges, Joseph Bondy-Denomy, Anna Maikova, Ekaterina Semenova, Konstantin Severinov, Olga Soutourina
mSphere,
2023
Single phage proteins sequester TIR- and cGAS-generated signaling molecules
Dong Li, Yu Xiao, Wijia Xiong, Iana Fedorova, Yu Wang, Xi Liu, Erin Huiting, Jie Ren, Zirui Gao, Xinguy Zhao, Xueli Coa, Yi Zhang, Joseph Bondy-Denomy, Yue Feng
bioRxiv,
2023
Anti-CRISPR proteins trigger a burst of CRISPR-Cas9 expression that enhances phage defense
Rachel E. Workman, Marie J. Stoltzfus, Nicholas C. Keith, Chad W. Euler, Joseph Bondy-Denomy, Joshua W. Modell
bioRxiv,
2023
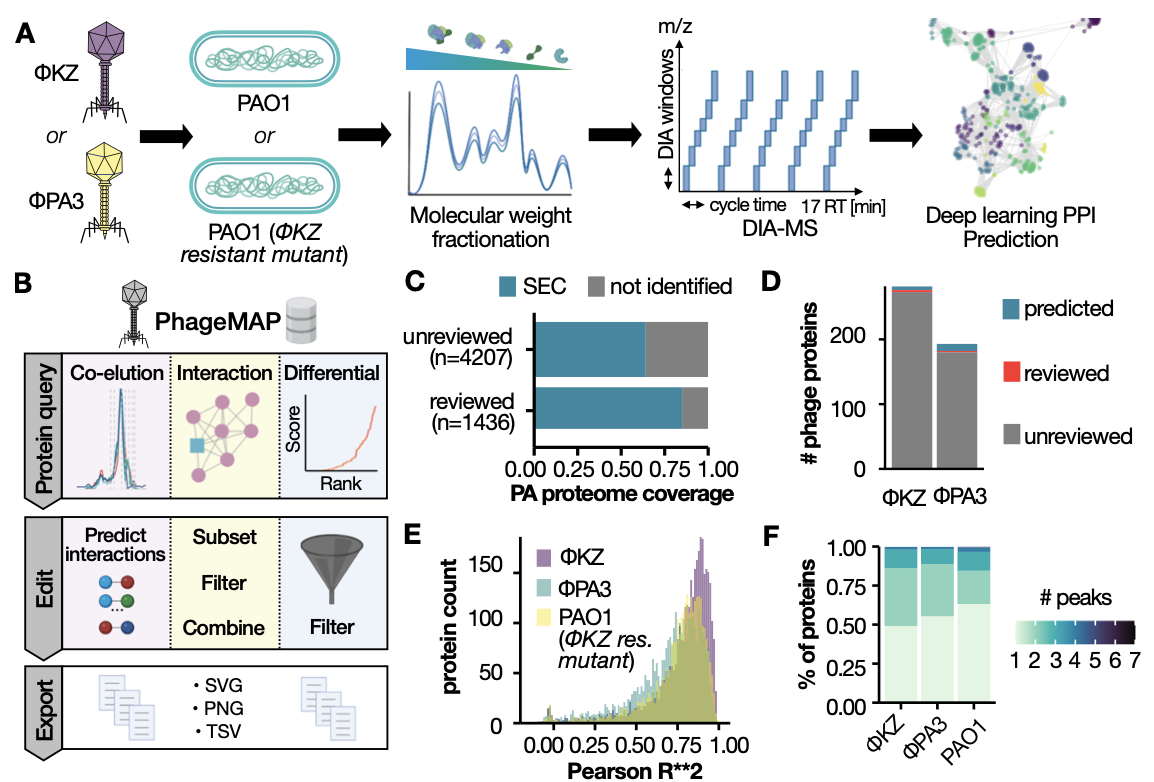
Next-generation interaction proteomics for quantitative Jumbophage-bacteria interaction mapping
Andrea Fossati, Deepto Mozumdar, Claire Kokontis, Eliza Nieweglowska, Melissa Mendez, Adrian Pelin, Yuping Li, Baron Guo, Nevan J. Krogan, David A. Agard, Joseph Bondy-Denomy and Danielle L. Swaney
Nature Communications,
2023
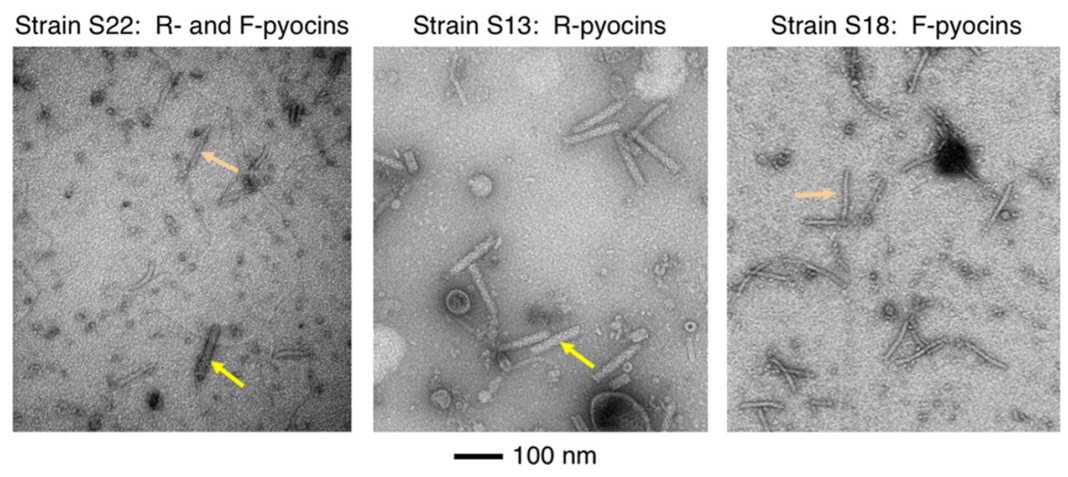
F-Type Pyocins Are Diverse Noncontractile Phage Tail-Like Weapons for Killing Pseudomonas aeruginosa
Senjuti Saha, Chidozie D. Ojobor, Annie Si Cong Li, Erik Mackinnon, Olesia I. North, Joseph Bondy-Denomy, Joseph S. Lam, Alexander W. Ensminger, Karen L. Maxwell, Alan R. Davidson
Journal of Bacteriology,
2023
Translation-dependent downregulation of Cas12a mR 1 NA by an anti-CRISPR protein
Nicole D. Marino, Alexander Talaie, Héloïse Carion, Matthew C. Johnson, Yang Zhang, Sukrit Silas, Yuping Li, and Joseph Bondy-Denomy
bioRxiv,
2023
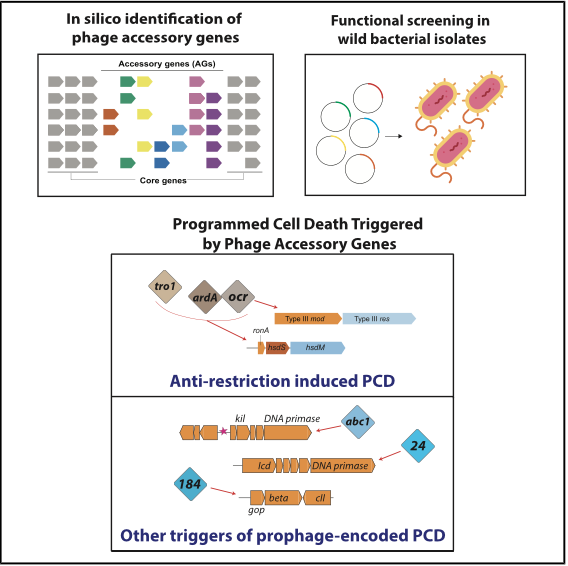
Activation of programmed cell death and counter-defense functions of phage accessory genes
Sukrit Silas, Héloïse Carion, Kira S. Makarova, Eric Laderman, David Sanchez Godinez, Matthew Johnson, Andrea Fossati, Danielle Swaney, Michael Bocek, Eugene V. Koonin, Joseph Bondy-Denomy
bioRxiv,
2023
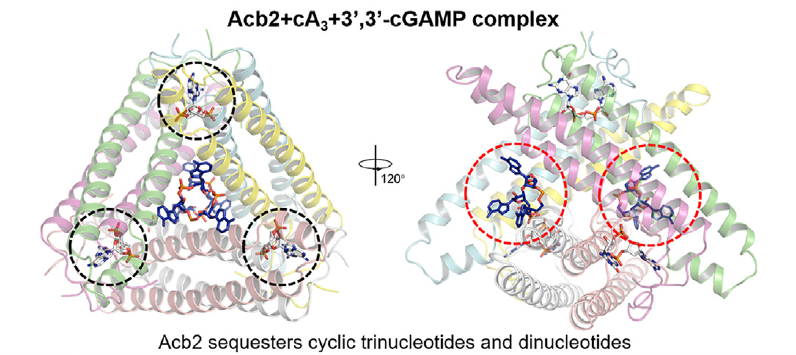
Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides
Xueli Cao, Yu Xiao, Erin Huiting, Xujun Cao, Dong Li, Jie Ren, Linlin Guan, Yu Wang, Lingyin Li, Joseph Bondy-Denomy, Yue Feng
Molecular Cell,
2023
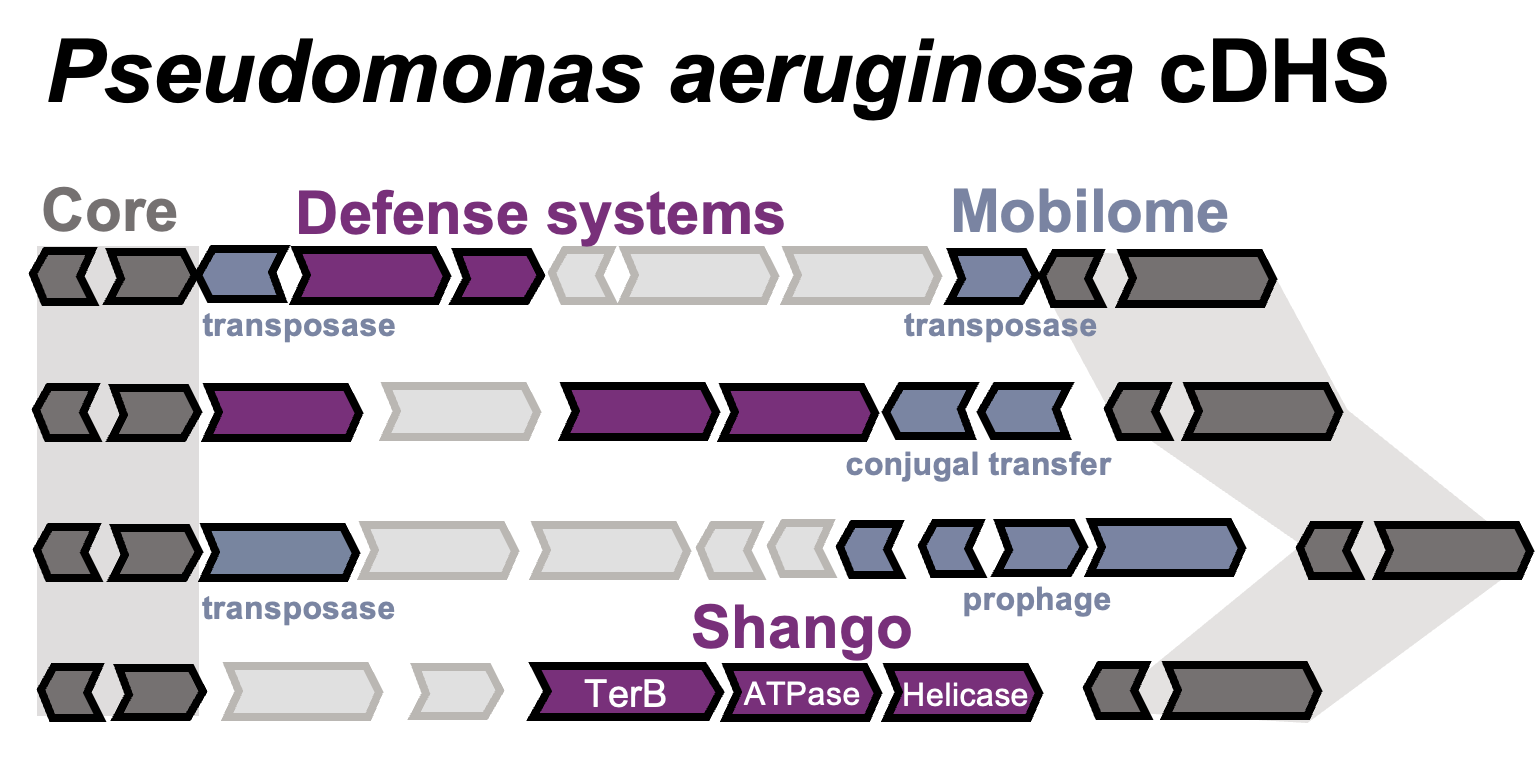
Core Defense Hotspots within Pseudomonas aeruginosa are a consistent and rich source of anti-phage defense systems
Matthew C. Johnson, Eric Laderman, Erin Huiting, Charles Zhang, Alan Davidson, Joseph Bondy-Denomy
Nucleic Acids Research,
2023
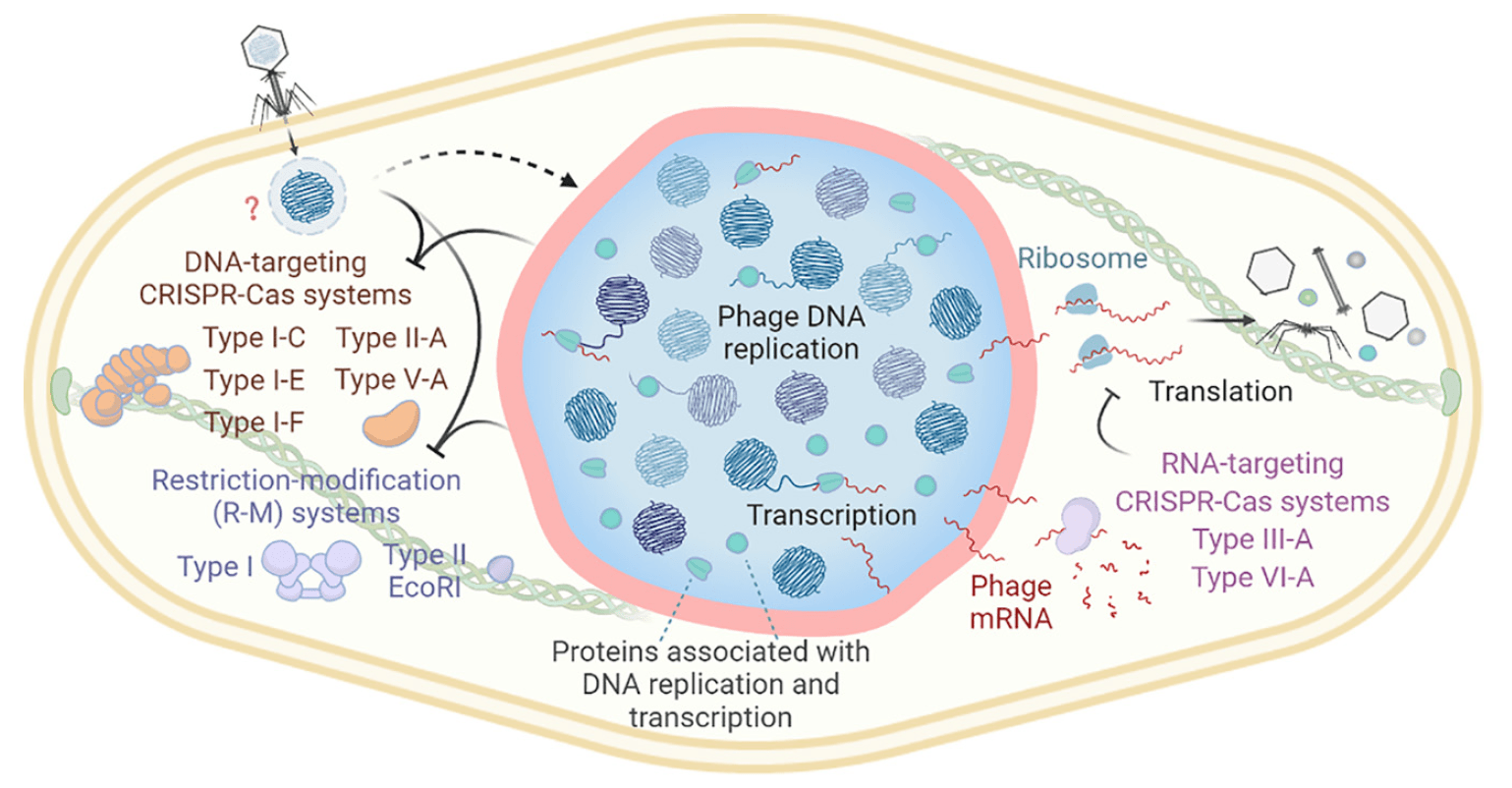
Defining the expanding mechanisms of phage-mediated activation of bacterial immunity
Erin Huiting, Joseph Bondy-Denomy
Current Opinion in Microbiology,
2023
Ten Years of Anti-CRISPR Research
Joseph Bondy-Denomy, Karen L Maxwell, Alan R Davidson
Journal of Molecular Biology,
2023
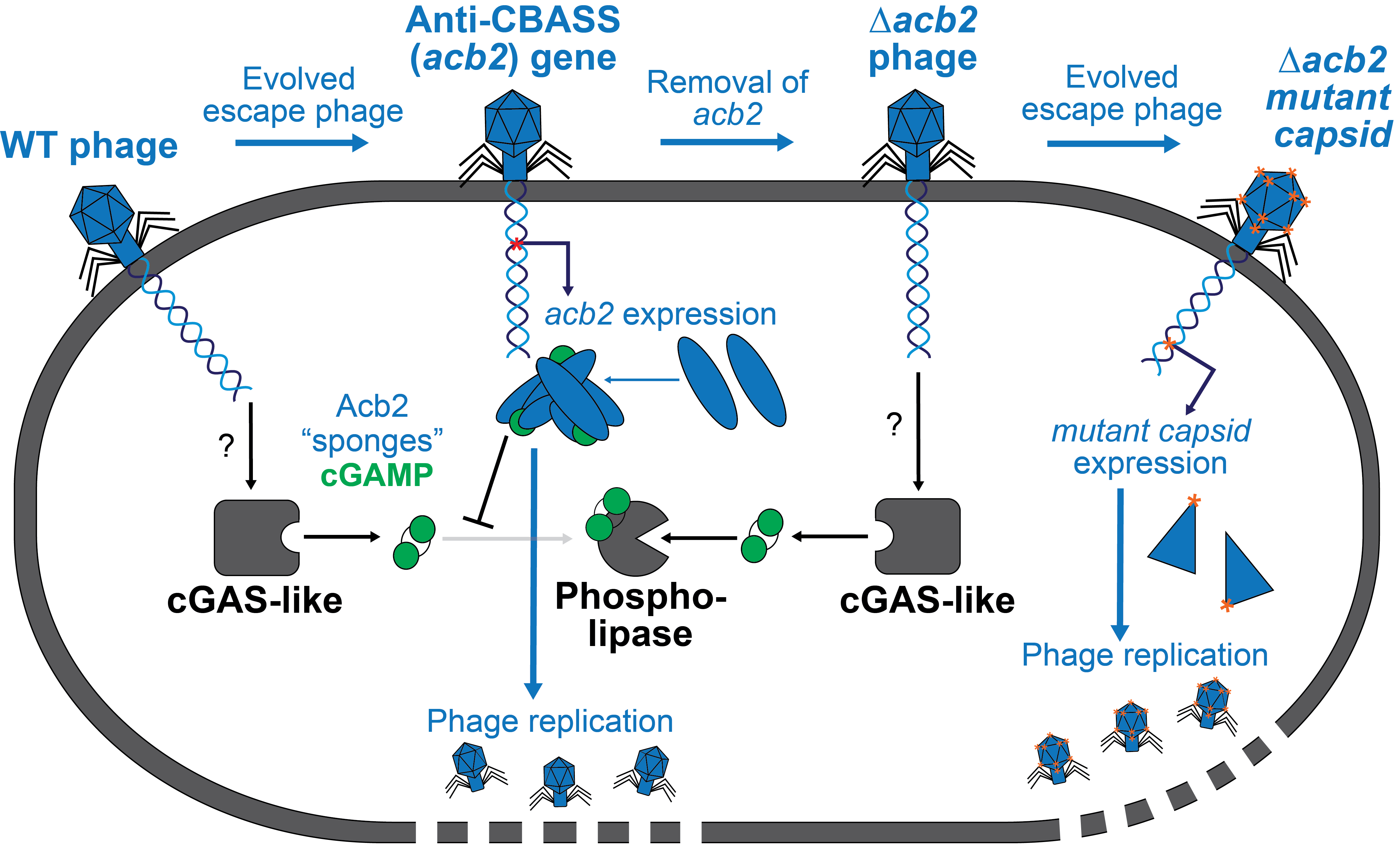
Bacteriophages inhibit and evade cGAS-like immune function in bacteria
Erin Huiting, Xueli Cao, Jie Ren, James S. Fraser, Yue Feng, Joseph Bondy-Denomy
Cell,
2023
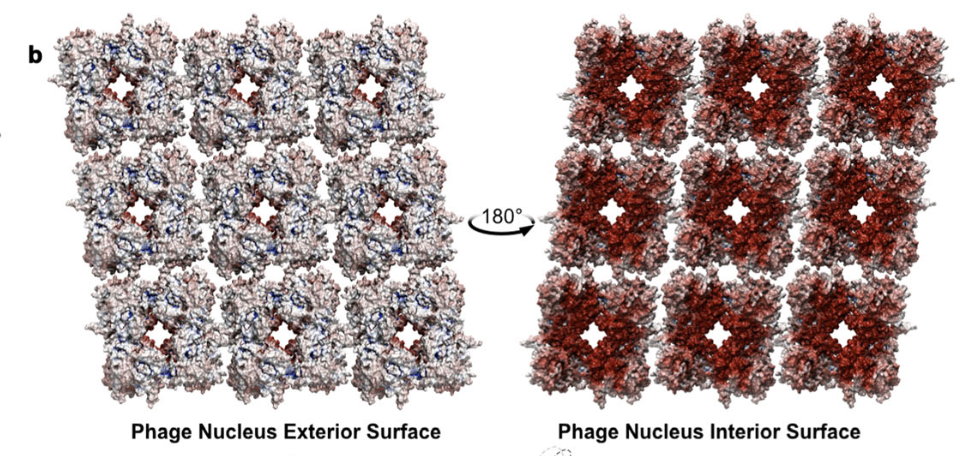
The φPA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice
Eliza S. Nieweglowska, Axel F. Brilot, Melissa Méndez-Moran, Claire Kokontis, Minkyung Baek, Junrui Li, Yifan Cheng, David Baker, Joseph Bondy-Denomy & David A. Agard
Nature Communications,
2023
Structural basis of AcrIF24 as an anti-CRISPR protein and transcriptional suppressor
Indranil Arun Mukherjee, Clinton Gabel, Nicholas Noinaj, Joseph Bondy-Denomy and Leifu Chang
Nature Chemical Biology,
2022
CRISPR-Cas12a targeting of ssDNA plays no detectable role in immunity
Nicole D. Marino, Rafael Pinilla-Redondo,and Joseph Bondy-Denomy
Nucleic Acids Research,
2022
Lack of Cas13a inhibition by anti-CRISPR proteins from Leptotrichia prophages
Matthew C. Johnson, Logan T. Hille, Benjamin P. Kleinstiver, Alexander J. Meeske, Joseph Bondy-Denomy
Molecular Cell,
2022
A single bacterial enzyme i(NHI)bits phage DNA replication
Erin Huiting and Joseph Bondy-Denomy
Cell Host and Microbe,
2022
Distinct Subcellular Localization of a Type I CRISPR Complex and the Cas3 Nuclease in Bacteria
Sutharsan Govindarajan, Adair Borges, Shweta Karambelkar, Joseph Bondy-Denomy
ASM Journal of Bacteriology,
2022
Bacteriophage genome engineering with CRISPR–Cas13a
Jingwen Guan, Agnès Oromí-Bosch, Senén D. Mendoza, Shweta Karambelkar, Joel D. Berry and Joseph Bondy-Denomy
Nature Microbiology,
2022
Genetic Manipulation of a CAST of Characters in a Microbial Community
Deepto Mozumdar Balint Csorgo and Joseph Bondy-Denomy**
The CRISPR Journal,
2022
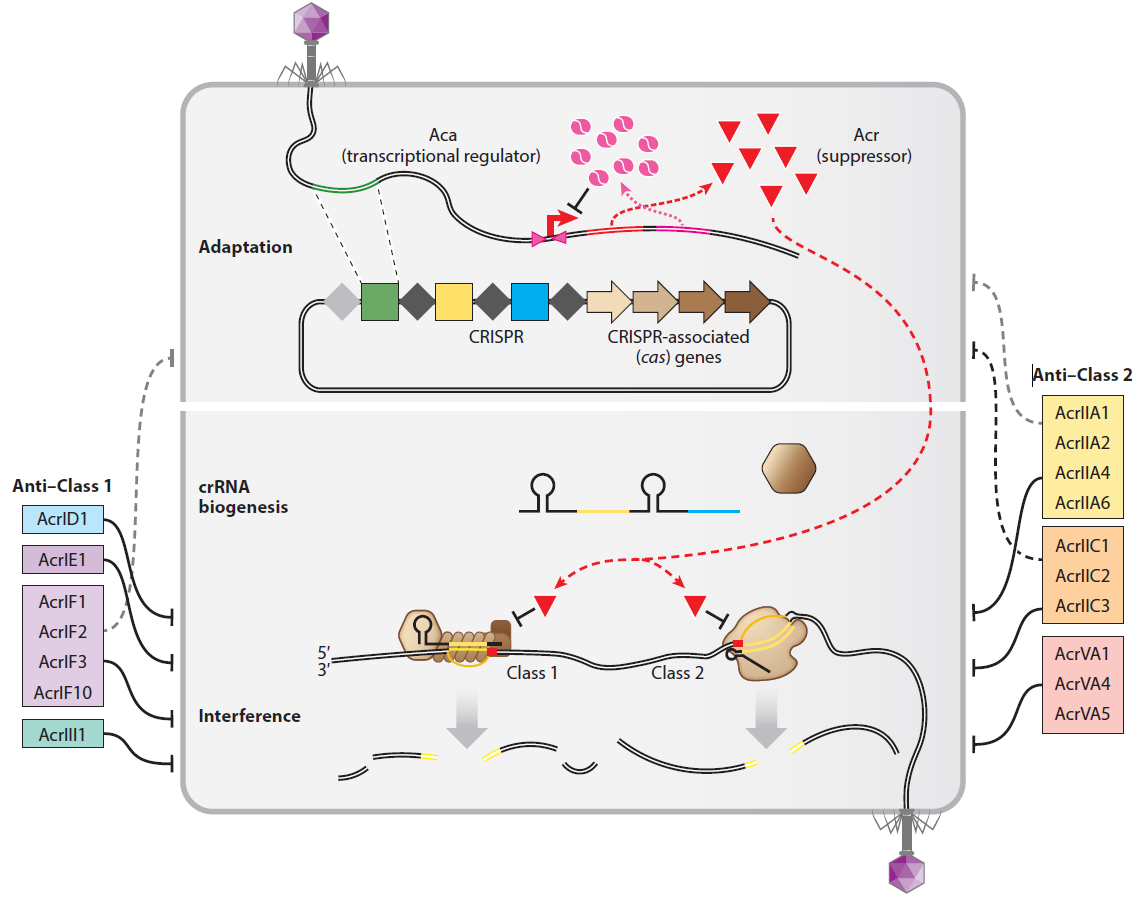
Anti-CRISPRs go viral: The infection biology of CRISPR-Cas inhibitors
Y Li, Bondy-Denomy J.
Cell Host and Microbe,
2021
Mobile element warfare via CRISPR and anti-CRISPR in Pseudomonas aeruginosa
León LM, Park AE, Borges AL, Zhang JY, Bondy-Denomy J.
Nucleic Acids Research,
2021
A phage-encoded anti-activator inhibits quorum sensing in Pseudomonas aeruginosa
Shah M, Taylor VL, Bona D, Tsao Y, Stanley SY, Pimentel-Elardo SM, McCallum M, Bondy-Denomy J, Howell PL, Nodwell JR, Davidson AR, Moraes TF, Maxwell KL.
Molecular Cell,
2021
Intracellular organization by jumbo bacteriophages
Guan J, Bondy-Denomy J.
Journal of Bacteriology,
2020
Discovery of multiple anti-CRISPRs highlights anti-defense gene clustering in mobile genetic elements
Pinilla-Redondo R, Shehreen S, Marino ND, Fagerlund ND, Brown CM, Sorensen SJ, Fineran PC, Bondy-Denomy J.
Nature Communications,
2020
A compact Cascade-Cas3 system for targeted genome engineering
Csörgő B, León LM, Chau-Ly IJ, Vasquez-Rifo A, Berry JD, Mahendra C, Crawford ED, Lewis JD, Bondy-Denomy J.
Nature methods,
2020
Machine-learning approach expands the repertoire of anti-CRISPR protein families
Gussow AB, Park AE, Borges AL, Shmakov SA, Makarova KS, Wolf YI, Bondy-Denomy J, Koonin EV.
Nature Communications,
2020
Listeria phages induce Cas9 degradation to protect lysogenic genomes
Osuna BA, Karambelkar S, Mahendra C, Christie KA, Garcia B, Davidson AR, Kleinstiver BP, Kilcher S, Bondy-Denomy J.
Cell Host & Microbe,
2020
Critical Anti-CRISPR locus repression by a bi-functional Cas9 inhibitor
Osuna BA, Karambelkar S, Mahendra C, Sarbach A, Johnson MC, Kilcher S, Bondy-Denomy J.
Cell Host & Microbe,
2020
Structures and strategies of anti-CRISPR-mediated immune suppression
Wiegand T, Karambelkar S, Bondy-Denomy J, Wiedenheft B.
Annual Reviews Microbiology,
2020
Broad-spectrum anti-CRISPR facilitate horizontal gene transfer
Mahendra C, Christie KA, Osuna BA, Pinilla-Redondo R, Kleinstiver BP, Bondy-Denomy J.
Nature Microbiololgy,
2020
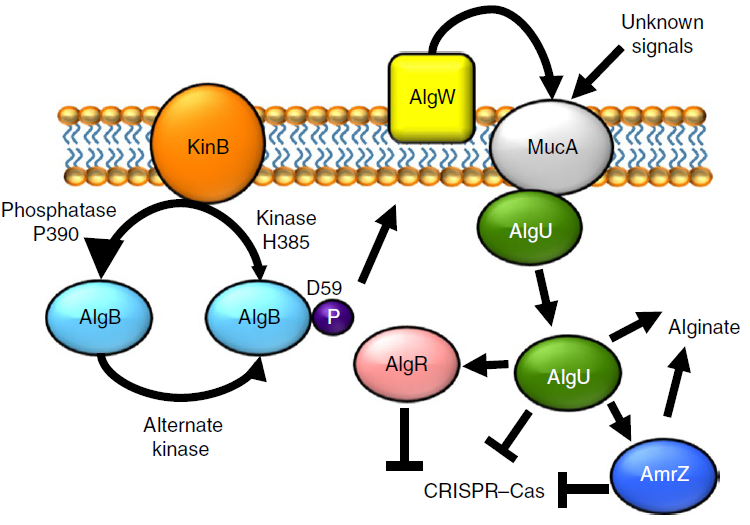
Bacterial alginate regulators and phage homologs repress CRISPR-Cas immunity
Borges AL, Castro B, Govindarajan S, Solvik T, Escalante V, Bondy-Denomy J.
Nature Microbiololgy,
2020
Anti-CRISPR protein applications: Natural breaks for CRISPR-Cas technologies
Marino N, Pinilla-Redondo R, Csörgő B, Bondy-Denomy J.
Nature Methods,
2020
A type IV-A CRISPR-Cas system in Pseudomonas aeruginosa mediates RNA-guided plasmid interference in vivo
Crowley VM, Catching A, Taylor HN, Borges AL, Metcalf J, Bondy-Denomy J, Jackson RN.
The CRISPR Journal,
2019
A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases
Mendoza S, Niewglowska ES, Govindarajan S, Leon LM, Berry JD, Tiwari A, Chaikeeratisak V, Pogliano J, Agard DA, Bondy-Denomy J.
Nature,
2019
Molecular mechanism of azithromycin resistance among typhoidal Salmonella strains in Bangladesh identified through passive pediatric surveillance
Hooda Y, Sajib MSI, Rahman H, Luby SP, Bondy-Denomy J, Santosham M, Andrews JR, Saha SK, Saha S.
PloS Neglected Tropical Diseases,
2019
CRISPR-Cas system of a prevalent human gut bacterium reveals hyper-targeting against phages in a human virome catalog
Soto-Perez P, Bisanz JE, Berry JD, Lam KN, Bondy-Denomy J, Turnbaugh PJ.
Cell Host & Microbe,
2019
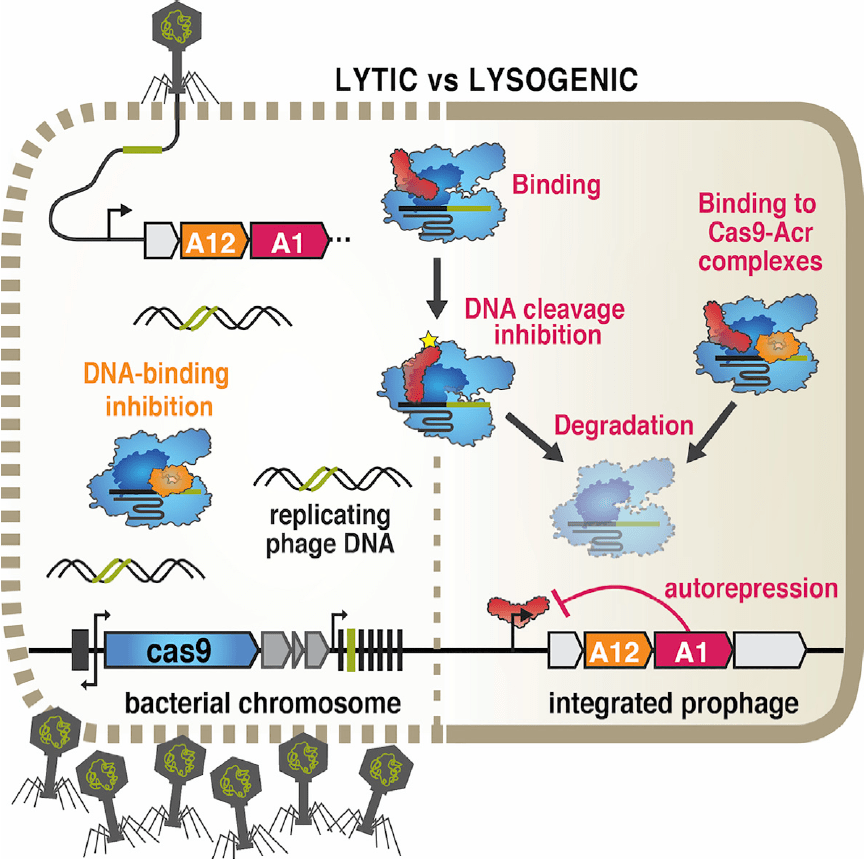
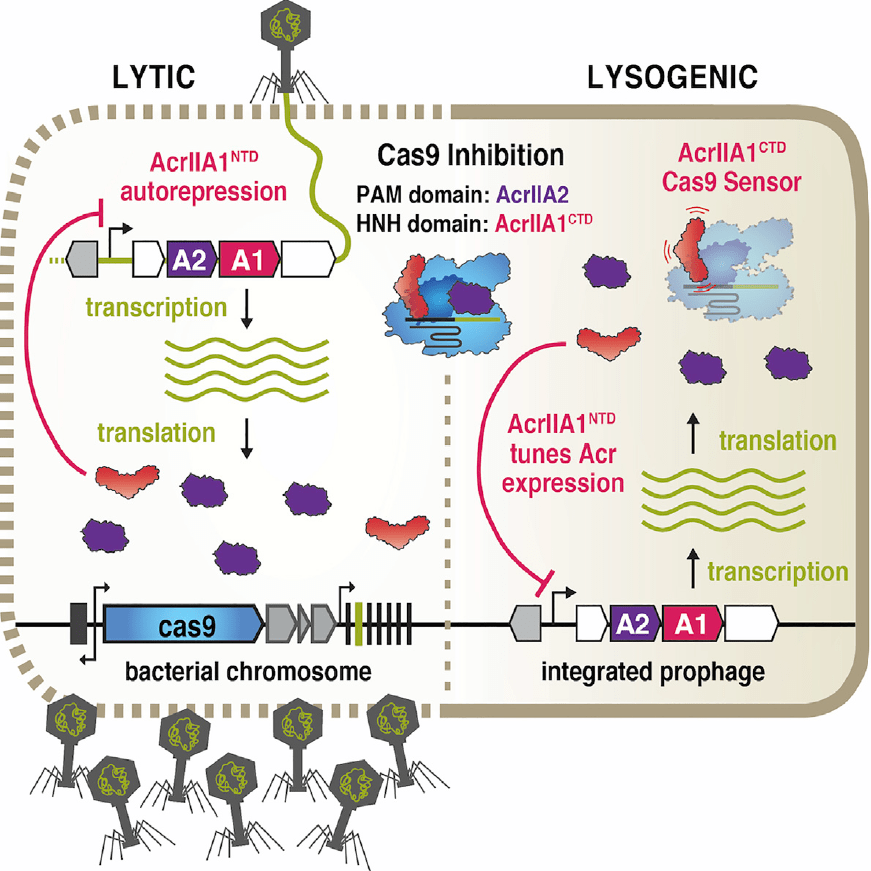
Anti-CRISPR-associated proteins are crucial repressors of anti-CRISPR transcription
Stanley SY, Borges AL, Chen KH, Swaney DL, Krogan NJ, Bondy-Denomy J, Davidson AR.
Cell,
2019
Cryptic inoviruses revealed as pervasive in bacteria and archaea in Earth’s biomes
Roux S, Krupovic M, Daly RA, Borges AL, Nayfach S, Schulz F, Sharrar A, Matheus Carnevali PB, Cheng JF, Ivanova NN, Bondy-Denomy J, Wrighton KC, Woyke T, Visel A, Kyrpides NC, Eloe-Fadrosh EA.
Nature Microbiology,
2019
Cas13 helps bacteria play dead when the enemy strikes
Mendoza SD, Bondy-Denomy J.
Cell Host & Microbe,
2019
Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells
Nakamura M, Srinivasan P, Chavez M, Carter MA, Dominguez AA, La Russa M, Lau MB, Abbott TR, Xu X, Zhao D, Gao Y, Kipniss NH, Smolke CD, Bondy-Denomy J, Qi LS.
Nature Communications,
2019
Temperature-responsive competitive inhibition of CRISPR-Cas9
Jiang F*, Liu JJ*, Osuna BA*, Xu M, Berry JD, Rauch BJ, Nogales E, Bondy-Denomy J, Doudna JA.
Nature Communications,
2018
Phage morons play an important role in Pseudomonas aeruginosa phenotypes
Tsao YF, Taylor VL, Kala S, Bondy-Denomy J, Khan AN, Bona D, Cattoir V, Lory S, Davidson AR, Maxwell KL.
Journal of Bacteriology,
2018
Discovery of widespread type I and type IV CRISPR-Cas inhibitors
Marino ND*, Zhang JY*, Borges AL*, Sousa AA, Leon LM, Rauch BJ, Walton RT, Berry JD, Joung JK, Kleinstiver BP, Bondy-Denomy J.
Science,
2018
A unified resource for tracking anti-CRISPR names
Bondy-Denomy J, Davidson AR, Doudna JA, Fineran PC, Maxwell KL, Moineau S, Peng X, Sontheimer EJ, Wiedenheft B.
The CRISPR Journal,
2018
Bacteriophage cooperation suppresses CRISPR-Cas3 and Cas9 immunity
Borges AL, Zhang JY, Rollins MF, Osuna BA, Wiedenheft B, Bondy-Denomy J.
Cell,
2018
Protein inhibitors of CRISPR-Cas9
Bondy-Denomy J.
ACS Chemical Biology,
2018
Pseudomonas aeruginosa defends against phages through type IV pilus glycosylation
Harvey H, Bondy-Denomy J, Marquis H, Sztanko KM, Davidson AR, Burrows LL.
Nature Microbiology,
2017
The Discovery, Mechanisms, and Evolutionary Impact of Anti-CRISPRs
Borges AL, Davidson AR, Bondy-Denomy J.
Annual Review of Virology,
2017
Disabling Cas9 by an anti-CRISPR DNA mimic
Shin J, Jiang F, Liu JJ, Bray NL, Rauch BJ, Baik SH, Nogales E, Bondy-Denomy J, Corn JE, Doudna JA.
Science Advances,
2017
Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity
Rollins MF, Chowdhury S, Carter J, Golden SM, Wilkinson RA, Bondy-Denomy J, Lander GC, Wiedenheft B.
PNAS,
2017
How bacteria control the CRISPR-Cas arsenal
Leon LM, Mendoza SD, Bondy-Denomy J.
Current Opinion in Microbiology,
2017
Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex
Chowdhury S, Carter J, Rollins MF, Golden SM, Jackson RN, Hoffmann C, Nosaka L, Bondy-Denomy J, Maxwell KL, Davidson AR, Fischer ER, Lander GC, Wiedenheft B.
Cell,
2017
CRISPR control of virulence in Pseudomonas aeruginosa
Wiedenheft B, Bondy-Denomy J.
Cell Research,
2017
Inhibition of CRISPR-Cas9 with bacteriophage proteins
Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, Bondy-Denomy J.
Cell,
2017
Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system
Høyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy-Denomy J., Bassler BL
PNAS,
2016
Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system
Høyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy-Denomy J., Bassler BL
Nature Communications,
2016
Prophages mediate defense against phage infection through diverse mechanisms
Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, Maxwell KL.
The ISME Journal,
2016
The diversity-generating benefits of a prokaryotic adaptive immune system
van Houte S, Ekroth AK, Broniewski JM, Chabas H, Ben Ashby, Bondy-Denomy J, Gandon S, Boots M, Paterson S, Buckling A, Westra ER.
Nature,
2016
Active and adaptive Legionella CRISPR-Cas reveals a recurrent challenge to the pathogen
Rao C, Guyard C, Pelaz C, Wasserscheid J, Bondy-Denomy J, Dewar K, Ensminger AW.
Cell Microbiology,
2016
Foreign DNA acquisition by the I-F CRISPR-Cas system requires all components of the interference machinery
Vorontsova D, Datsenko KA, Medvedeva S, Bondy-Denomy J, Savitskaya EE, Pougach K, Logacheva M, Wiedenheft B, Davidson AR, Severinov K, Semenova E.
Nucleic Acids Research,
2015
Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins
Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, Davidson AR.
Nature,
2015
Parasite exposure drives selective evolution of constitutive verses inducible defense
Westra ER, van Houte S, Oyesiku-Blakemore S, Makin B, Broniewski JM, Best A, Bondy-Denomy J, Davidson A, Boots M, Buckling A.
Current Biology,
2015
A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa
Pawluk A, Bondy-Denomy J, Cheung VH, Maxwell KL, Davidson AR.
mBio,
2014
To acquire or resist: The complex biological effects of CRISPR-Cas systems
Bondy-Denomy J, Davidson AR.
Trends in Microbiology,
2014
When a virus is not a parasite: The beneficial effects of prophages on bacterial fitness
Bondy-Denomy J, Davidson AR.
Journal of Microbiology,
2014
Efficacy of bacteriophage treatment on Pseudomonas aeruginosa biofilms
Phee A, Bondy-Denomy J, Kishen A, Basrani B, Azarpazhooh A, Maxwell K.
Journal of Endodontics,
2013
Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system
Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR.
Nature,
2013
The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages
Cady KC*, Bondy-Denomy J*, Heussler GE, Davidson AR, O’Toole GA.
Journal of Bacteriology,
2012
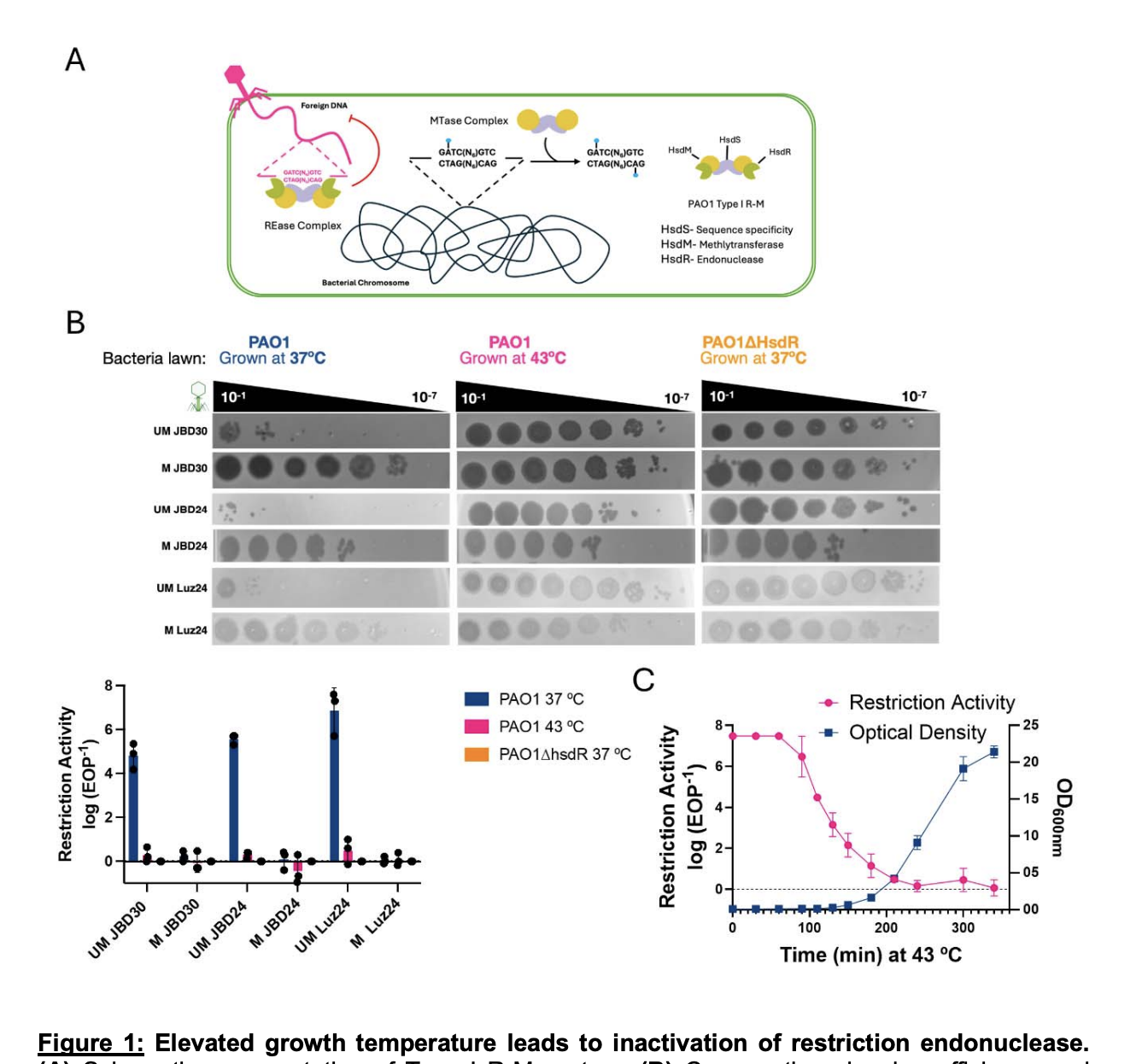
Multigenerational Proteolytic Inactivation of Restriction Upon Subtle Genomic Hypomethylation
Shmidov E, Villani A, Mendoza SD, Avihu E, Lebenthal-Loinger I, Karako-Lampert S, Shoshani S, Ye C, Wang Y, Yan H, Tang W, Bondy-Denomy J, Banin E.
bioRxiv,
10